Identification of protein binding sites on U3 snoRNA and pre-rRNA by UV cross-linking and high-throughput analysis of cDNAs - PubMed (original) (raw)
Identification of protein binding sites on U3 snoRNA and pre-rRNA by UV cross-linking and high-throughput analysis of cDNAs
Sander Granneman et al. Proc Natl Acad Sci U S A. 2009.
Abstract
The U3 small nucleolar ribonucleoprotein (snoRNP) plays an essential role in ribosome biogenesis but, like many RNA-protein complexes, its architecture is poorly understood. To address this problem, binding sites for the snoRNP proteins Nop1, Nop56, Nop58, and Rrp9 were mapped by UV cross-linking and analysis of cDNAs. Cross-linked protein-RNA complexes were purified under highly-denaturing conditions, ensuring that only direct interactions were detected. Recovered RNA fragments were amplified after linker ligation and cDNA synthesis. Cross-linking was successfully performed either in vitro on purified complexes or in vivo in living cells. Cross-linking sites were precisely mapped either by Sanger sequencing of multiple cloned fragments or direct, high-throughput Solexa sequencing. Analysis of RNAs associated with the snoRNP proteins revealed remarkably high signal-to-noise ratios and identified specific binding sites for each of these proteins on the U3 RNA. The results were consistent with previous data, demonstrating the reliability of the method, but also provided insights into the architecture of the U3 snoRNP. The snoRNP proteins were also cross-linked to pre-rRNA fragments, with preferential association at known sites of box C/D snoRNA function. This finding demonstrates that the snoRNP proteins directly contact the pre-rRNA substrate, suggesting roles in snoRNA recruitment. The techniques reported here should be widely applicable to analyses of RNA-protein interactions.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Fig. 1.
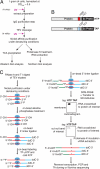
The CRAC technique. (A) Purification of protein–RNA complexes. Cells were UV-irradiated in Petri dishes on ice. Extracts were incubated with IgG beads and tagged proteins were released by TEV protease cleavage. Cross-linked complexes were purified via nickel affinity purification under denaturing conditions. Purified proteins were detected by Western analysis and cross-linked RNAs were detected by Northern analysis. (B) Schematic representation of a protein fused to either the HTP tag (Upper) or the TAP tag (Lower). Prot A: Staphylococcus aureus Protein A IgG binding domain. (C) Identification of RNA binding sites. Partially Rnase-digested RNPs were incubated with nickel beads to immobilize His6-tagged proteins (blue ovals) and covalently attached RNAs (red lines). Cross-linked RNAs were 3′ dephosphorylated, ligated to the adenylated linker (blue line), radioactively-labeled with polynucleotide kinase, and then ligated to the 5′ linker (green line). After release by imidazole treatment, radioactive RNPs were resolved on Bis-Tris NuPAGE gels and transferred to nitrocellulose. Bands corresponding to the predicted _M_r of the target protein were excised and digested with proteinase K, and recovered RNAs were amplified by RT/PCR. The PCR products were gel-purified and sequenced. ddC: dideoxy-cytidine. InvddT: inverted dideoxythymidine. The asterisk indicates the UV cross-linking site.
Fig. 2.
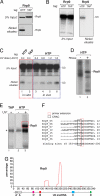
Mapping Rrp9 cross-linking sites. (A) Rrp9-HTP is specifically recovered on nickel beads. Extracts from cells expressing HTP or TAP-tagged Rrp9 were purified as in Fig. 1_A_. Five percent of the TEV eluates (5% Input) and the nickel eluates (Nickel eluates) were resolved on 4–12% Bis-Tris NuPAGE gels and detected by Western analysis. (B) Rrp9-HTP is cross-linked to U3. RNA extracted from 2% of the TEV eluates (2% Input) and nickel eluates (Nickel eluates) was analyzed by Northern analysis. (C) Rrp9 UV cross-links to the U3 snoRNA in vivo. Rrp9-HTP (lanes 1 and 3–6) or Rrp9-TAP (lane 2) were purified as shown in Fig. 1_C_. UV cross-linking was performed in vitro (lanes 1 and 2) or in vivo (lanes 3–6) and cross-linked U3 was detected by Northern analysis. The UV dose (J/cm2) is indicated above each lane. (D) Protein is cross-linked to radiolabeled RNA. CRAC was performed with strains expressing Rrp9-HTP with (lane 2) or without (lane 1) RNase treatment. (E) Contaminant proteins are not associated with RNA. CRAC was performed with strains expressing Rrp9-HTP with (lane 2) or without (lane 1) UV cross-linking. The asterisk indicates frequently-detected contaminants. Dashed red boxes indicate regions from which cross-linked RNA was extracted. (F) Multiple sequence alignment for the major Rrp9 binding sites. The black box indicates where deletions were frequently identified. The dashed red box indicates 2 primer extension stops detected after cross-linking (
Fig. S1
). (G) Histogram displaying locations of Rrp9-associated RNA fragments mapped to the U3 snoRNA (x axis). Percentage (y axis) is the number of reads mapped to that nucleotide divided by the total of U3 reads.
Fig. 3.
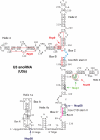
Overview of U3 CRAC results. Schematic representation of the architecture of the U3 snoRNP complex. Colored nucleotides indicate conserved boxes B (red), C (blue), C′ (pink), and D (green). Box A and A′ are marked by black boxes. The open circles indicate nucleotides that were frequently mutated in cross-linking experiments. Arrows point to predicted cross-linking sites of the individual proteins. The dashed boxes indicate the conserved stem II at the box B/C and box C′/D motif.
Fig. 4.
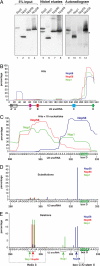
CRAC identifies U3-binding sites of box C/D snoRNP proteins. (A) HTP-tagged proteins were cross-linked and purified from cell extracts as in Fig. 1_A_. Five percent of the TEV eluates (5% Input) and the nickel eluates were resolved on 4–12% Bis-Tris NuPAGE gels and detected by Western blot analysis (lanes 1–8) or autoradiography (lanes 9–12). The asterisk indicates the location of the contaminant ≈55-kDa band. (B) Binding sites of common box C/D snoRNP proteins to the U3 snoRNA. Nop1 (green), Nop56 (red), and Nop58 (blue) all primarily bind the 3′ end of U3. The histogram represents all sequences mapped to the U3 snoRNA, irrespective of length. Percentage (y axis) was calculated as in Fig. 2. Locations of functionally-important elements in U3 are indicated. (C) Nop1 (green), Nop56 (red), and Nop58 (blue) bind U3 at distinct sites. Locations of hits <18 nt in the 3′ region of U3 (nucleotides 290–333) are shown. Percentages are the numbers of short reads mapped to that nucleotide divided by the total of short U3 reads in this region. U3 sequence coordinates are indicated on the x axis. The green box indicates the box D sequence. (D and E) Distribution of substitutions (D) and deletions (E) in U3 sequences cross-linked to Nop1 (green), Nop56 (red), and Nop58 (blue). Percentage is the frequency of nucleotide substitutions or deletions, divided by the total number of reads at that site. Brackets indicate regions where mutations were most frequently identified. Sequences of the stem II of the box C/D motif and helix 3 are indicated by dashed boxes. Arrows point to predicted protein cross-linking sites.
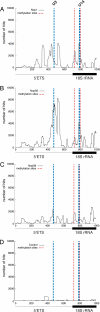
Fig. 5.
snoRNP proteins directly bind the pre-rRNA. (A–C) Binding sites for Nop1, Nop56, and Nop58 across the 5′ region of the pre-rRNA. (D) CRAC results for the untagged control strain. Red lines indicate sites of snoRNA-directed 2′-O-methylation. Blue lines indicate the site U3 base-pairing in the 5′ETS and the U14 base-pairing in the 18S rRNA, which are required for pre-rRNA processing. Hits on the complete 35S pre-rRNA region are presented in
Figs. S2 and S3
.
Similar articles
- Implication of the box C/D snoRNP assembly factor Rsa1p in U3 snoRNP assembly.
Rothé B, Manival X, Rolland N, Charron C, Senty-Ségault V, Branlant C, Charpentier B. Rothé B, et al. Nucleic Acids Res. 2017 Jul 7;45(12):7455-7473. doi: 10.1093/nar/gkx424. Nucleic Acids Res. 2017. PMID: 28505348 Free PMC article. - A pre-ribosomal RNA interaction network involving snoRNAs and the Rok1 helicase.
Martin R, Hackert P, Ruprecht M, Simm S, Brüning L, Mirus O, Sloan KE, Kudla G, Schleiff E, Bohnsack MT. Martin R, et al. RNA. 2014 Aug;20(8):1173-82. doi: 10.1261/rna.044669.114. Epub 2014 Jun 19. RNA. 2014. PMID: 24947498 Free PMC article. - Identification of protein factors and U3 snoRNAs from a Brassica oleracea RNP complex involved in the processing of pre-rRNA.
Samaha H, Delorme V, Pontvianne F, Cooke R, Delalande F, Van Dorsselaer A, Echeverria M, Sáez-Vásquez J. Samaha H, et al. Plant J. 2010 Feb 1;61(3):383-98. doi: 10.1111/j.1365-313X.2009.04061.x. Epub 2009 Nov 3. Plant J. 2010. PMID: 19891704 - The box C/D and H/ACA snoRNPs: key players in the modification, processing and the dynamic folding of ribosomal RNA.
Watkins NJ, Bohnsack MT. Watkins NJ, et al. Wiley Interdiscip Rev RNA. 2012 May-Jun;3(3):397-414. doi: 10.1002/wrna.117. Epub 2011 Nov 7. Wiley Interdiscip Rev RNA. 2012. PMID: 22065625 Review. - Mpp10p, a new protein component of the U3 snoRNP required for processing of 18S rRNA precursors.
Baserga SJ, Agentis TM, Wormsley S, Dunbar DA, Lee S. Baserga SJ, et al. Nucleic Acids Symp Ser. 1997;(36):64-7. Nucleic Acids Symp Ser. 1997. PMID: 9478208 Review.
Cited by
- The Efg1-Bud22 dimer associates with the U14 snoRNP contacting the 5' rRNA domain of an early 90S pre-ribosomal particle.
Beine-Golovchuk O, Kallas M, Kunze R, Griesel S, Baßler J. Beine-Golovchuk O, et al. Nucleic Acids Res. 2024 Jan 11;52(1):431-447. doi: 10.1093/nar/gkad1109. Nucleic Acids Res. 2024. PMID: 38000371 Free PMC article. - Pervasive transcription fine-tunes replication origin activity.
Candelli T, Gros J, Libri D. Candelli T, et al. Elife. 2018 Dec 17;7:e40802. doi: 10.7554/eLife.40802. Elife. 2018. PMID: 30556807 Free PMC article. - Contribution of domain structure to the function of the yeast DEDD family exoribonuclease and RNase T functional homolog, Rex1.
Daniels PW, Hama Soor T, Levicky Q, Hettema EH, Mitchell P. Daniels PW, et al. RNA. 2022 Apr;28(4):493-507. doi: 10.1261/rna.078939.121. Epub 2022 Jan 26. RNA. 2022. PMID: 35082142 Free PMC article. - HITS-CLIP and integrative modeling define the Rbfox splicing-regulatory network linked to brain development and autism.
Weyn-Vanhentenryck SM, Mele A, Yan Q, Sun S, Farny N, Zhang Z, Xue C, Herre M, Silver PA, Zhang MQ, Krainer AR, Darnell RB, Zhang C. Weyn-Vanhentenryck SM, et al. Cell Rep. 2014 Mar 27;6(6):1139-1152. doi: 10.1016/j.celrep.2014.02.005. Epub 2014 Mar 6. Cell Rep. 2014. PMID: 24613350 Free PMC article. - A quantitative analysis of CLIP methods for identifying binding sites of RNA-binding proteins.
Kishore S, Jaskiewicz L, Burger L, Hausser J, Khorshid M, Zavolan M. Kishore S, et al. Nat Methods. 2011 May 15;8(7):559-64. doi: 10.1038/nmeth.1608. Nat Methods. 2011. PMID: 21572407
References
- Gilbert C, Kristjuhan A, Winkler GS, Svejstrup JQ. Elongator interactions with nascent mRNA revealed by RNA immunoprecipitation. Mol Cell. 2004;14:457–464. - PubMed
- Gilbert C, Svejstrup JQ. Ausubel FM, editor. RNA immunoprecipitation for determining RNA-protein associations in vivo. Current Protocols in Molecular Biology. 2006:27.4.1–27.4.11. Chapter 27. - PubMed
- Urlaub H, Hartmuth K, Kostka S, Grelle G, Luhrmann R. A general approach for identification of RNA-protein cross-linking sites within native human spliceosomal small nuclear ribonucleoproteins (snRNPs). Analysis of RNA-protein contacts in native U1 and U4/U6.U5 snRNPs. J Biol Chem. 2000;275:41458–41468. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- G0900740/MRC_/Medical Research Council/United Kingdom
- WT_/Wellcome Trust/United Kingdom
- BB/D019621/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases