Discrimination of common and unique RNA-binding activities among Fragile X mental retardation protein paralogs - PubMed (original) (raw)
. 2009 Sep 1;18(17):3164-77.
doi: 10.1093/hmg/ddp255. Epub 2009 Jun 1.
Affiliations
- PMID: 19487368
- PMCID: PMC2722981
- DOI: 10.1093/hmg/ddp255
Discrimination of common and unique RNA-binding activities among Fragile X mental retardation protein paralogs
Jennifer C Darnell et al. Hum Mol Genet. 2009.
Abstract
Fragile X mental retardation is caused by loss-of-function of a single gene encoding FMRP, an RNA-binding protein that harbors three canonical RNA-binding domains, two KH-type and one RGG box. Two autosomal paralogs of FMRP, FXR1P and FXR2P, are similar to FMRP in their overall structure, including the presence of putative RNA-binding domains, but to what extent they provide functional redundancy with FMRP is unclear. Although FMRP has been characterized as a polyribosome-associated regulator of translation, less is known about the functions of FXR1P and FXR2P. For example, FMRP binds intramolecular G-quadruplex and kissing complex RNA (kcRNA) ligands via the RGG box and KH2 domain, respectively, although the RNA ligands of FXR1P and FXR2P are unknown. Here we demonstrate that FXR1P and FXR2P KH2 domains bind kcRNA ligands with the same affinity as the FMRP KH2 domain although other KH domains do not. RNA ligand recognition by this family is highly conserved, as the KH2 domain of the single Drosophila ortholog, dFMRP, also binds kcRNA. kcRNA was able to displace FXR1P and FXR2P from polyribosomes as it does for FMRP, and this displacement was FMRP-independent. This suggests that all three family members recognize the same binding site on RNA mediating their polyribosome association, and that they may be functionally redundant with regard to this aspect of translational control. In contrast, FMRP is unique in its ability to recognize G-quadruplexes, suggesting the FMRP RGG domain may play a non-redundant role in the pathophysiology of the disease.
Figures
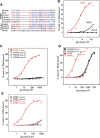
Figure 1.
The KH2 domain variable loop encoded by exons 11 and 12 (E11 and E12), absent from FXR1P and FXR2P, does not contribute to kcRNA binding. (A) Diagram depicting the sequences present in the recombinant human KH2 domains that were produced in bacteria and purified. Domains differed in the presence or absence of exons 11 and 12 and in the presence of the I304N mutation in the ‘parent’ construct as a negative control for kcRNA binding. (B) Filter binding assays were performed to determine the affinity of the indicated domains in (A) for binding to the kc2 RNA. Domains were tested for RNA binding immediately after purification. (C) Mouse (m, squares) and human (h, circles) KH2 domains with and without the alternatively spliced exon 12 (filled symbols, +E12; open symbols, -E12) were assayed for binding to kcRNA by filter binding assay. Although kcRNA binding is conserved between mouse and human, the presence of exon 12 sequences inhibits binding. In this experiment, domains were tested for RNA binding following storage at 4°C (see text for further explanation of the observed differences in binding for the constructs containing E12 in B and C).
Figure 2.
KH2 domains from human (h), mouse (m) and Drosophila (d) FXR family members are conserved in key positions. Signature alpha helices (red boxes), beta sheets (blue boxes) and loops (green boxes) are shown. The asterisks denote hydrophobic amino acids whose side chains make up the aliphatic α/β platform of the domain. Red dots denote amino acids important for interactions of human Nova-2 KH3 with its RNA ligand. An open triangle indicates the isoleucine mutated to asparagine in the I304N patient, and the dashed gray line denotes the exon–exon junction between exons 10 and 13 of FMRP, with exons 11 and 12 removed, or exons 10 and 11 of FXR1P and FXR2P. In the dFMRP sequence, number 3 represents nonaligned amino acids ‘AIA’, and number 2, ‘NI’. RNA contacts from the Nova2 KH3:RNA co-crystal structures are shown below the sequences (8,44).
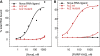
Figure 3.
The KH domains of Nova-1 and FMRP bind different RNA ligands. Human Nova-1 protein was expressed and HisTag-purified from a bacterial lysate. Filter binding assays were performed using Nova-1 and the human FMRP KH2 domain at the indicated concentrations with one of Nova's in vivo targets, the GABA-Aγ2 intron 9-binding site (46) (black squares, Nova ligand), or the 96 nucleotide _in vitro-_selected RNAs, kc2 (filled red squares, kc2wt) or the C50G point mutant in kc2 (half-filled red squares, kc2 mutant). (A) Nova bound its cognate ligand with an affinity of 37.1 n
m
(±16.0 n
m
). Binding to kc2wt or kc2 mutant was too low to determine a _K_d value. (B) FMRP KH2 bound its kcRNA ligand with _K_d = 37.8 ± 5.6 n
m
. Binding to kc2 mutant or the Nova ligand was not quantifiable.
Figure 4.
KH2 domains from human FMRP, FXR1P, FXR2P and dFMRP bind kcRNA specifically and indistinguishably. (A) FMRP KH2 (blue circles), FXR1P KH2 (green squares) or FXR2P KH2 (red triangles) were produced in bacteria and purified. Equilibrium filter binding assays with 32P-labeled kcRNA were used to determine the affinity of the KH2 domains for kc2 RNA ligand (_K_d value for FMRP = 43 n
m
, FXR1P = 42 n
m
and FXR2P = 60 n
m
, solid lines). Assays were repeated with the C50G mutant kcRNA (dashed lines) indicating the specificity of the FXRP interaction with this RNA. (B) The KH2 domains from human (open squares) and Drosophila (closed squares) were expressed and purified side-by-side and kcRNA binding assessed by filter binding assay as in (A).
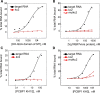
Figure 5.
The KH domains of Sf1, Vg1RBP/Vera or PCBP1 bind their previously identified RNA ligands, but do not bind with significant affinity to kcRNA or mutant kcRNA. (A) The isolated KH-QUA domain of human Sf1 bound the intronic branch point sequence with _K_d = 2.83 ± 0.71 µ
m
(black squares). (B) All four KH domains of Xenopus Vera bound to the Vg1 RNA (black squares) with _K_d = 292 ± 80 n
m
. (C) Binding of the mouse PCBP1 tandem KH1 and KH2 domains to the R7a1 ligand (black squares) had a _K_d value of 42.3 ± 12 n
m
. (D) Binding of the KH3 domain of PCBP1 to the R7a1 ligand (black squares) had a _K_d value of 2.17 ± 0.25 µ
m
. In all cases, binding to kc2 RNA (red squares) or C50G mutant kc2RNA (half-filled red squares) was too low to accurately determine a _K_d value.
Figure 6.
FMRP, FXR1P and FXR2P use the same mechanism to associate with polysomes. (A) Postmitochondrial supernatants prepared from cortex and cerebellum of Fmr1 null mice were treated with buffer (control), 500 n
m
kc2 (kc2) or 500 n
m
mutant kc2 (mut kc2) containing a point mutation that disrupts the loop–loop interaction in the kissing motif. Lysates were then fractionated over linear 20–50% sucrose gradients, proteins recovered from each fraction by TCA precipitation and resolved by SDS–PAGE. FXR1P or FXR2P was visualized by western blot with ML13 or 1G2 antibodies respectively. (B) kcRNA treatment has no effect on overall ribosome distribution on transcripts as evidenced by A254 traces.
Figure 7.
Several polysome-associated proteins are not competed off polysomes by kcRNA treatment. (A) Postmitochondrial supernatants prepared from the cortex of WT P21 mice were treated with buffer (control), with 500 n
m
kc2 (kc2), or 500 n
m
mutant kc2 (mut kc2). Lysates were then fractionated over linear 20–50% sucrose gradients, proteins recovered from each fraction by TCA precipitation and resolved by SDS–PAGE. Half as much lysate was precipitated for lanes 1–3 as for 4–16 to permit full solubilization of large TCA pellets. Proteins were visualized by western blot with antibodies as detailed in Materials and Methods. (B) kcRNA and mutant kcRNA treatment have no effect on the polysome profile as evidenced by A254 traces.
Figure 8.
G-quadruplex binding by the RGG box is specific to FMRP. (A) Alignment of FXR family RGG box sequences from the indicated species by ClustalW analysis. (B) The C-termini of human FMRP (red circles), FXR1P (black squares) and FXR2P (black triangles) were assayed for binding to sc1 by filter binding assay. (C) Fusion proteins corresponding to sequences encoded by exons 12 (open squares), 13 (circles) or 14 (triangles) of the FXR2P C-terminus were assayed for binding to sc1 RNA compared with the full-length C-terminus of FMRP (closed red squares). (D) Filter binding assay of sc1 binding by fusion protein corresponding to exons 15–17 of FXR2P (black, _K_d = 1.05 ± 0.18 µ
m
in K+) and the C-terminus of FMRP (red, _K_d = 6.5 ± 1.9 n
m
in K+) was conducted in buffer containing 200 m
m
K+ (closed squares) or 200 m
m
Li+ (open squares). (E) The C-terminus of dFMRP (black) was assayed for sc1 (closed) or mutant sc1 (open) binding compared with the C-terminus of human FMRP (red) by filter binding assay.
Similar articles
- Kissing complex RNAs mediate interaction between the Fragile-X mental retardation protein KH2 domain and brain polyribosomes.
Darnell JC, Fraser CE, Mostovetsky O, Stefani G, Jones TA, Eddy SR, Darnell RB. Darnell JC, et al. Genes Dev. 2005 Apr 15;19(8):903-18. doi: 10.1101/gad.1276805. Epub 2005 Apr 1. Genes Dev. 2005. PMID: 15805463 Free PMC article. - RNA-Binding Specificity of the Human Fragile X Mental Retardation Protein.
Athar YM, Joseph S. Athar YM, et al. J Mol Biol. 2020 Jun 12;432(13):3851-3868. doi: 10.1016/j.jmb.2020.04.021. Epub 2020 Apr 25. J Mol Biol. 2020. PMID: 32343993 Free PMC article. - Interactions of the G quartet forming semaphorin 3F RNA with the RGG box domain of the fragile X protein family.
Menon L, Mihailescu MR. Menon L, et al. Nucleic Acids Res. 2007;35(16):5379-92. doi: 10.1093/nar/gkm581. Epub 2007 Aug 9. Nucleic Acids Res. 2007. PMID: 17693432 Free PMC article. - FMRP RNA targets: identification and validation.
Darnell JC, Mostovetsky O, Darnell RB. Darnell JC, et al. Genes Brain Behav. 2005 Aug;4(6):341-9. doi: 10.1111/j.1601-183X.2005.00144.x. Genes Brain Behav. 2005. PMID: 16098133 Review. - Biology of the fragile X mental retardation protein, an RNA-binding protein.
Khandjian EW. Khandjian EW. Biochem Cell Biol. 1999;77(4):331-42. Biochem Cell Biol. 1999. PMID: 10546896 Review.
Cited by
- FXR1 associates with and degrades PDZK1IP1 and ATOH8 mRNAs and promotes esophageal cancer progression.
Khan FA, Fouad D, Ataya FS, Fang N, Dong J, Ji S. Khan FA, et al. Biol Direct. 2024 Nov 7;19(1):104. doi: 10.1186/s13062-024-00553-3. Biol Direct. 2024. PMID: 39511680 Free PMC article. - FMRP cooperates with miRISC components to repress translation and regulate neurite morphogenesis in Drosophila.
Kaul N, Pradhan SJ, Boin NG, Mason MM, Rosales J, Starke EL, Wilkinson EC, Chapman EG, Barbee SA. Kaul N, et al. RNA Biol. 2024 Jan;21(1):11-22. doi: 10.1080/15476286.2024.2392304. Epub 2024 Aug 27. RNA Biol. 2024. PMID: 39190491 Free PMC article. - An in-depth review of the function of RNA-binding protein FXR1 in neurodevelopment.
Méndez-Albelo NM, Sandoval SO, Xu Z, Zhao X. Méndez-Albelo NM, et al. Cell Tissue Res. 2024 Nov;398(2):63-77. doi: 10.1007/s00441-024-03912-8. Epub 2024 Aug 19. Cell Tissue Res. 2024. PMID: 39155323 Review. - PRMT5-mediated arginine methylation of FXR1 is essential for RNA binding in cancer cells.
Vijayakumar A, Majumder M, Yin S, Brobbey C, Karam J, Howley B, Howe PH, Berto S, Madan LK, Gan W, Palanisamy V. Vijayakumar A, et al. Nucleic Acids Res. 2024 Jul 8;52(12):7225-7244. doi: 10.1093/nar/gkae319. Nucleic Acids Res. 2024. PMID: 38709899 Free PMC article. - FMRP-mediated spatial regulation of physiologic NMD targets in neuronal cells.
Kurosaki T, Rambout X, Maquat LE. Kurosaki T, et al. Genome Biol. 2024 Jan 23;25(1):31. doi: 10.1186/s13059-023-03146-x. Genome Biol. 2024. PMID: 38263082 Free PMC article. Review.
References
- Gibson T.J., Rice P.M., Thompson J.D., Heringa J. KH domains within the FMR1 sequence suggest that fragile X syndrome stems from a defect in RNA metabolism. Trends Biochem. Sci. 1993;18:331–333. - PubMed
- Ashley C.T., Wilkinson K.D., Reines D., Warren S.T. FMR-1 protein: conserved RNP family domains and selective RNA binding. Science. 1993;262:563–566. - PubMed
- Siomi H., Siomi M.C., Nussbaum R.L., Dreyfuss G. The protein product of the fragile X gene, FMR1, has characteristics of an RNA-binding protein. Cell. 1993;74:291–298. - PubMed
- DeBoulle K., Verkerk A.J., Reyniers E., Vits L., Hendrickx J., Van Roy B., Van Den Bos F., de Graaff E., Oostra B.A., Willems P.J. A point mutation in the FMR-1 gene associated with fragile X mental retardation. Nat. Genet. 1993;3:31–35. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- 5R01 HD40647/HD/NICHD NIH HHS/United States
- NS40955/NS/NINDS NIH HHS/United States
- R01 HD040647/HD/NICHD NIH HHS/United States
- R01 HD40647/HD/NICHD NIH HHS/United States
- R01S NS34389/NS/NINDS NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases