Genetic modifiers of dFMR1 encode RNA granule components in Drosophila - PubMed (original) (raw)
Genetic modifiers of dFMR1 encode RNA granule components in Drosophila
Anne-Marie J Cziko et al. Genetics. 2009 Aug.
Abstract
Mechanisms of neuronal mRNA localization and translation are of considerable biological interest. Spatially regulated mRNA translation contributes to cell-fate decisions and axon guidance during development, as well as to long-term synaptic plasticity in adulthood. The Fragile-X Mental Retardation protein (FMRP/dFMR1) is one of the best-studied neuronal translational control molecules and here we describe the identification and early characterization of proteins likely to function in the dFMR1 pathway. Induction of the dFMR1 in sevenless-expressing cells of the Drosophila eye causes a disorganized (rough) eye through a mechanism that requires residues necessary for dFMR1/FMRP's translational repressor function. Several mutations in dco, orb2, pAbp, rm62, and smD3 genes dominantly suppress the sev-dfmr1 rough-eye phenotype, suggesting that they are required for dFMR1-mediated processes. The encoded proteins localize to dFMR1-containing neuronal mRNPs in neurites of cultured neurons, and/or have an effect on dendritic branching predicted for bona fide neuronal translational repressors. Genetic mosaic analyses indicate that dco, orb2, rm62, smD3, and dfmr1 are dispensable for translational repression of hid, a microRNA target gene, known to be repressed in wing discs by the bantam miRNA. Thus, the encoded proteins may function as miRNA- and/or mRNA-specific translational regulators in vivo.
Figures
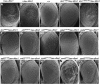
Figure 1.—
Scanning electron micrographs of Drosophila compound eyes demonstrating suppression of sev-dfmr1 induced rough eyes by mutations in discs overgrown/doubletime (dco), orb2, poly-A binding protein (pabp), smd3, and rm62/dmp68. Two representative EMS or _P_-element associated alleles were selected with the median phenotypes and where possible, a revertant of a representative insertional allele isolated by _P_-element mobilization. A sev-dfmr1 transgene allows dFMR1 protein to be overexpressed in a subset of photoreceptors via the sevenless (sev) promoter. (A and B) Eyes of control animals. F1 progeny from a cross between sev-dfmr1 and w1118 flies showing the range of rough-eye phenotypes. (C) Wild-type (Canton S) eyes. (D) The _P_-element-associated dcoS139602 allele in trans with sev-dfmr1. (E) Eyes of phenotypic “revertant” dcoS139602-REV/sev-dfmr1 flies show reversion of suppression following _P_-element excision from dcoS139602. (F) dcoS05813/sev-dfmr1. (G and H, respectively) orb2BG02373/sev-dfmr1 and orb2delta2/sev-dfmr1. (I and J, respectively) pabpK10109/sev-dfmr1 and pabpEP310/sev-dfmr1. (K and L, respectively) rm6201086/sev-dfmr1 and rm62EY06795/sev-dfmr1. (M) smd3EP2176/sev-dfmr1 and (N) phenotypic revertant of smd3EP2176. Eyes of smd3EP2176-REV/sev-dfmr1 flies: smd3EP2176-REV was isolated by the mobilization and excision of the EP2176 P-insertion. (O) smd3K09029/sev-dfmr1.
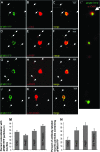
Figure 2.—
Orb2, poly-A binding protein (PABP), Rm62/Dmp68, and Sm proteins are present on dFMR1-containing foci in neurites of cultured Drosophila primary neurons. Neurites of dissociated and cultured larval neurons contain distinct FMR1-containing mRNPs that also contain several molecules involved in translational control (B
arbee
et al. 2006; B
eckham
et al. 2008; K
wak
et al. 2008). These particles visualized in processes of motor neuron expressing dFMR1-YFP (from C380Gal4; ChaGal80; UASdfmr1-YFP larvae) also contain Orb2 (A–C), PABP (D–F), Rm62 (G–I), and Sm (J–L) proteins. C′ shows an expanded and higher-resolution image of the boxed region in C to clearly demonstrate the observed colocalization. The percentage of colocalization between dFMR1YFP and the other proteins in neuritic granules is quantified in M (which shows the fraction of dFMR1-YFP marked neuritic particles that also contain each tested protein) and N (which indicates the fraction of neurite granules with each tested protein that also contains dFMR1-YFP). The signal in somata necessarily appears overexposed, to clearly image fainter neuritic granules that are the focus of our colocalization analysis.
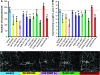
Figure 3.—
Overexpression of identified proteins reduces dendritic branching and alters dendritic morphology in larval class IV sensory neurons. Each of the sev-dfmr1 interacting genes was overexpressed via UAS/Gal4 technology in sensory neurons using a flp-out technique, in which Gal4477 driver was combined with an act<CD2<Gal4, UAS Flip recombinase, to mark fine dendritic processes (B
arbee
et al. 2006). (A) Number of branches/cells was decreased in all lines overexpressing Discs overgrown/Doubletime (dcoEP3280, dcoEY02910), poly-A binding protein lines (UAS-pabp (L), UAS pabp (B), pabpEP310, pabpEY11561), or Rm62 (rm62EP3607, rm62EY01915, rm62 EY06975) and one of the two SmD3 (smD3EP2104) lines assayed. (B) Total dendrite length was significantly reduced in all poly-A binding protein lines (UAS-pabp (L), UAS pabp (B), pabpEP310, pabpEY11561), all Discs overgrown/Doubletime (dcoEP3280, dcoEY02910), two Rm62 (rm62EP3607, rm62EY0191), and one of the SmD3 (smD3EP2104) lines assayed. We do not have a definitive explanation why smd3EP2176 does not show a similar effect. The dashed line indicates control levels. (C) Representative images of a labeled sensory neuron from each overexpression line are presented.
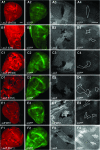
Figure 4.—
Suppressors of sev-dfmr1 as well as the dfmr1 gene are not essential for efficient _bantam-_miRNA mediated repression of a target translational reporter (hid reporter GFP). (A–F) show analyses of _hid_-reporter expression in mutant clones of smd3 (A), dco/dbt (B), rm62 (C), orb2 (D), dfmr1(E), and dicer-1 (F). (A–F) panel 1, 20× magnification images of mutant clones in wing imaginal discs visualized by staining with a corresponding antibody or LacZ; panel 2, 20× GFP-labeled hid reporter expression, whose levels are low due to efficient _bantam_-mediated translational repression; panel 3, 60× close-up of mutant clones marked with corresponding antibody or LacZ; and panel 4, corresponding area of clones (circled) with _hid_-GFP expression. (F), 1–4, Dicer1 clones show significant upregulation of the hid reporter. See
methods
for larval genotypes.
Similar articles
- FMRP and Ataxin-2 function together in long-term olfactory habituation and neuronal translational control.
Sudhakaran IP, Hillebrand J, Dervan A, Das S, Holohan EE, Hülsmeier J, Sarov M, Parker R, VijayRaghavan K, Ramaswami M. Sudhakaran IP, et al. Proc Natl Acad Sci U S A. 2014 Jan 7;111(1):E99-E108. doi: 10.1073/pnas.1309543111. Epub 2013 Dec 16. Proc Natl Acad Sci U S A. 2014. PMID: 24344294 Free PMC article. - Fragile X mental retardation protein modulates the fate of germline stem cells in Drosophila.
Yang L, Duan R, Chen D, Wang J, Chen D, Jin P. Yang L, et al. Hum Mol Genet. 2007 Aug 1;16(15):1814-20. doi: 10.1093/hmg/ddm129. Epub 2007 May 21. Hum Mol Genet. 2007. PMID: 17519221 - Genetic and systems level analysis of Drosophila sticky/citron kinase and dFmr1 mutants reveals common regulation of genetic networks.
Bauer CR, Epstein AM, Sweeney SJ, Zarnescu DC, Bosco G. Bauer CR, et al. BMC Syst Biol. 2008 Nov 25;2:101. doi: 10.1186/1752-0509-2-101. BMC Syst Biol. 2008. PMID: 19032789 Free PMC article. - RNA interference: a new mechanism by which FMRP acts in the normal brain? What can Drosophila teach us?
Siomi H, Ishizuka A, Siomi MC. Siomi H, et al. Ment Retard Dev Disabil Res Rev. 2004;10(1):68-74. doi: 10.1002/mrdd.20011. Ment Retard Dev Disabil Res Rev. 2004. PMID: 14994291 Review. - dFmr1 Plays Roles in Small RNA Pathways of Drosophila melanogaster.
Specchia V, D'Attis S, Puricella A, Bozzetti MP. Specchia V, et al. Int J Mol Sci. 2017 May 16;18(5):1066. doi: 10.3390/ijms18051066. Int J Mol Sci. 2017. PMID: 28509881 Free PMC article. Review.
Cited by
- Modeling Fragile X Syndrome in Drosophila.
Drozd M, Bardoni B, Capovilla M. Drozd M, et al. Front Mol Neurosci. 2018 Apr 16;11:124. doi: 10.3389/fnmol.2018.00124. eCollection 2018. Front Mol Neurosci. 2018. PMID: 29713264 Free PMC article. Review. - Drosophila CPEB Orb2A mediates memory independent of Its RNA-binding domain.
Krüttner S, Stepien B, Noordermeer JN, Mommaas MA, Mechtler K, Dickson BJ, Keleman K. Krüttner S, et al. Neuron. 2012 Oct 18;76(2):383-95. doi: 10.1016/j.neuron.2012.08.028. Epub 2012 Oct 17. Neuron. 2012. PMID: 23083740 Free PMC article. - The RNA-binding proteins FMR1, rasputin and caprin act together with the UBA protein lingerer to restrict tissue growth in Drosophila melanogaster.
Baumgartner R, Stocker H, Hafen E. Baumgartner R, et al. PLoS Genet. 2013;9(7):e1003598. doi: 10.1371/journal.pgen.1003598. Epub 2013 Jul 11. PLoS Genet. 2013. PMID: 23874212 Free PMC article. - Drosophila modeling of heritable neurodevelopmental disorders.
Gatto CL, Broadie K. Gatto CL, et al. Curr Opin Neurobiol. 2011 Dec;21(6):834-41. doi: 10.1016/j.conb.2011.04.009. Epub 2011 May 17. Curr Opin Neurobiol. 2011. PMID: 21596554 Free PMC article. Review. - Genes and pathways affected by CAG-repeat RNA-based toxicity in Drosophila.
Shieh SY, Bonini NM. Shieh SY, et al. Hum Mol Genet. 2011 Dec 15;20(24):4810-21. doi: 10.1093/hmg/ddr420. Epub 2011 Sep 20. Hum Mol Genet. 2011. PMID: 21933837 Free PMC article.
References
- Aakalu, G., W. B. Smith, N. Nguyen, C. Jiang and E. M. Schuman, 2001. Dynamic visualization of local protein synthesis in hippocampal neurons. Neuron 30 489–502. - PubMed
- Ashraf, S. I., A. L. McLoon, S. M. Sclarsic and S. Kunes, 2006. Synaptic protein synthesis associated with memory is regulated by the RISC pathway in Drosophila. Cell 124 191–205. - PubMed
- Barbee, S. A., A. L. Lublin and T. C. Evans, 2002. A novel function for the Sm proteins in germ granule localization during C. elegans embryogenesis. Curr. Biol. 12 1502–1506. - PubMed
- Barkoff, A. F., K. S. Dickson, N. K. Gray and M. Wickens, 2000. Translational control of cyclin B1 mRNA during meiotic maturation: coordinated repression and cytoplasmic polyadenylation. Dev. Biol. 220 97–109. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases