Molecular interactions in rotavirus assembly and uncoating seen by high-resolution cryo-EM - PubMed (original) (raw)
Molecular interactions in rotavirus assembly and uncoating seen by high-resolution cryo-EM
James Z Chen et al. Proc Natl Acad Sci U S A. 2009.
Abstract
Rotaviruses, major causes of childhood gastroenteritis, are nonenveloped, icosahedral particles with double-strand RNA genomes. By the use of electron cryomicroscopy and single-particle reconstruction, we have visualized a rotavirus particle comprising the inner capsid coated with the trimeric outer-layer protein, VP7, at a resolution (4 A) comparable with that of X-ray crystallography. We have traced the VP7 polypeptide chain, including parts not seen in its X-ray crystal structure. The 3 well-ordered, 30-residue, N-terminal "arms" of each VP7 trimer grip the underlying trimer of VP6, an inner-capsid protein. Structural differences between free and particle-bound VP7 and between free and VP7-coated inner capsids may regulate mRNA transcription and release. The Ca(2+)-stabilized VP7 intratrimer contact region, which presents important neutralizing epitopes, is unaltered upon capsid binding.
Conflict of interest statement
Conflict of interest statement: E.C.S. and P.R.D. are employees and shareholders of Novartis Vaccines and Diagnostics, Inc.
Figures
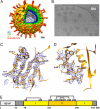
Fig. 1.
Rotavirus structure determined by cryo-EM. (A) Structure of the complete virion filtered at 25-Å resolution. The segmentation of the structure is based on reconstructions of the complete virion and the VP7-coated DLP and on published work of others (–23). (B) Cryo-EM image of VP7-coated DLPs (7RP). (C and D) Regions of the VP7 density (filtered to 4.2-Å resolution) corresponding to strands β2, β1, β3, and β11 and helix αB. (E) Domain organization of VP7 (17). Domain I (light yellow) is a Rossmann fold; domain II (dark yellow), a β-jelly roll. The N- and C-terminal extensions (gray) become ordered when VP7 associates with the DLP. Numbers below the bar are those of the first residue in each segment. Disulfide bonds are indicated as residue numbers joined by lines across the top of the bar. The signal peptide is 50 residues long. Numbering corresponds to rhesus rotavirus, serotype G3.
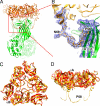
Fig. 2.
VP7 outer protein layer. (A) Structure of a VP6–VP7 heterohexamer, derived from crystal structures of VP6 (green) (14) and VP7 (gold) (16), docked into the cryo-EM density, and from the model built into cryo-EM density for the N-terminal arm. Two of the three VP7 N termini forming tight interactions with VP6 can be seen on the left and right side of the VP6 trimer. The red square indicates the area shown in more detail in B. (B) VP7 N terminus (residues 58–78) and corresponding cryo-EM density, showing its interaction with VP6. Pro-58 and Asn-69 are labeled; the latter bears a glycan for which density is present. Strands A′ and A″ of VP6 are also labeled. The density corresponding to residues 61–68 is weaker than in other parts of the map because this part of the structure lacks a clearly defined secondary structure and makes no contact with other parts of the structure. The map was filtered at 4.5-Å resolution to display a continuous density trace in this region. (C and D) Conformational differences between bound VP7 (gold) and the VP7 crystal structure (red) (16). The VP7 trimer is viewed from the outside of the virus along its symmetry axis (C) and from the “side”, normal to its symmetry axis (D). The way in which the domain hinge displacement “flattens” the subunit when the trimer binds VP6 is evident in the latter view. The alignment is based on a superposition of the Rossmann-fold domains.
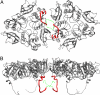
Fig. 3.
Interface between 2 VP7 trimers. The interface is shown from the outside of the virus (A) and from the “side”, normal to its symmetry axes (B). The N-terminal arms between residues 58 and 78 are shown as red curves; the approximate positions of the arms between residues 51 (the N terminus) and 57 are shown as dotted green curves, illustrating that the N termini may contribute to intertrimer contacts.
Fig. 4.
Conformational differences between DLP and 7RP. The structures of VP2 and VP6 docked into the 7RP cryo-EM density are shown in green whereas the DLP structure (12) is shown in red. A narrowing of the central channel on the icosahedral 5-fold axis and an inward movement of the VP2 and VP6 layers in the 7RP, compared with the DLP, are the principal differences. The VP7 layer of the 7RP is not shown (for clarity); the green arrow indicates the “clamping down” of the VP6 and VP2 layers that accompanies VP7 binding.
Comment in
- Single-particle cryoEM reconstructions: meeting the challenge.
Rey FA. Rey FA. Proc Natl Acad Sci U S A. 2009 Jun 30;106(26):10398-9. doi: 10.1073/pnas.0905001106. Epub 2009 Jun 24. Proc Natl Acad Sci U S A. 2009. PMID: 19553214 Free PMC article. No abstract available.
Similar articles
- Single-particle cryoEM reconstructions: meeting the challenge.
Rey FA. Rey FA. Proc Natl Acad Sci U S A. 2009 Jun 30;106(26):10398-9. doi: 10.1073/pnas.0905001106. Epub 2009 Jun 24. Proc Natl Acad Sci U S A. 2009. PMID: 19553214 Free PMC article. No abstract available. - Projection structure of VP6, the rotavirus inner capsid protein, and comparison with bluetongue VP7.
Hsu GG, Bellamy AR, Yeager M. Hsu GG, et al. J Mol Biol. 1997 Sep 26;272(3):362-8. doi: 10.1006/jmbi.1997.1179. J Mol Biol. 1997. PMID: 9325096 - Atomic structure of the major capsid protein of rotavirus: implications for the architecture of the virion.
Mathieu M, Petitpas I, Navaza J, Lepault J, Kohli E, Pothier P, Prasad BV, Cohen J, Rey FA. Mathieu M, et al. EMBO J. 2001 Apr 2;20(7):1485-97. doi: 10.1093/emboj/20.7.1485. EMBO J. 2001. PMID: 11285213 Free PMC article. - Rotavirus Replication: Gaps of Knowledge on Virus Entry and Morphogenesis.
Suzuki H. Suzuki H. Tohoku J Exp Med. 2019 Aug;248(4):285-296. doi: 10.1620/tjem.248.285. Tohoku J Exp Med. 2019. PMID: 31447474 Review. - Structural studies on orbivirus proteins and particles.
Stuart DI, Grimes JM. Stuart DI, et al. Curr Top Microbiol Immunol. 2006;309:221-44. doi: 10.1007/3-540-30773-7_8. Curr Top Microbiol Immunol. 2006. PMID: 16909901 Review.
Cited by
- On the effect of thermodynamic equilibrium on the assembly efficiency of complex multi-layered virus-like particles (VLP): the case of rotavirus VLP.
Roldão A, Mellado MC, Lima JC, Carrondo MJ, Alves PM, Oliveira R. Roldão A, et al. PLoS Comput Biol. 2012;8(2):e1002367. doi: 10.1371/journal.pcbi.1002367. Epub 2012 Feb 16. PLoS Comput Biol. 2012. PMID: 22359487 Free PMC article. - Fabs enable single particle cryoEM studies of small proteins.
Wu S, Avila-Sakar A, Kim J, Booth DS, Greenberg CH, Rossi A, Liao M, Li X, Alian A, Griner SL, Juge N, Yu Y, Mergel CM, Chaparro-Riggers J, Strop P, Tampé R, Edwards RH, Stroud RM, Craik CS, Cheng Y. Wu S, et al. Structure. 2012 Apr 4;20(4):582-92. doi: 10.1016/j.str.2012.02.017. Epub 2012 Apr 3. Structure. 2012. PMID: 22483106 Free PMC article. - Interactions among capsid proteins orchestrate rotavirus particle functions.
Trask SD, Ogden KM, Patton JT. Trask SD, et al. Curr Opin Virol. 2012 Aug;2(4):373-9. doi: 10.1016/j.coviro.2012.04.005. Epub 2012 May 16. Curr Opin Virol. 2012. PMID: 22595300 Free PMC article. Review. - Generation of single-round infectious rotavirus with a mutation in the intermediate capsid protein VP6.
Kotaki T, Kanai Y, Onishi M, Minami S, Chen Z, Nouda R, Nurdin JA, Yamasaki M, Kobayashi T. Kotaki T, et al. J Virol. 2024 Jul 23;98(7):e0076224. doi: 10.1128/jvi.00762-24. Epub 2024 Jun 5. J Virol. 2024. PMID: 38837379 Free PMC article. - Fusion of the mouse IgG1 Fc domain to the VHH fragment (ARP1) enhances protection in a mouse model of rotavirus.
Günaydın G, Yu S, Gräslund T, Hammarström L, Marcotte H. Günaydın G, et al. Sci Rep. 2016 Jul 21;6:30171. doi: 10.1038/srep30171. Sci Rep. 2016. PMID: 27439689 Free PMC article.
References
- Estes MK, Kapikian AZ. Rotaviruses. In: Howley DMKPM, editor. Fields Virology. 5th ed. Philadelphia: Lippincott, Williams & Wilkins; 2007. pp. 1918–1974.
- Adams WR, Kraft LM. Epizootic diarrhea of infant mice: Indentification of the etiologic agent. Science. 1963;141:359–360. - PubMed
- Altenburg BC, Graham DY, Estes MK. Ultrastructural study of rotavirus replication in cultured cells. J Gen Virol. 1980;46:75–85. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- AI-053174/AI/NIAID NIH HHS/United States
- P01 GM062580/GM/NIGMS NIH HHS/United States
- R01 AI053174/AI/NIAID NIH HHS/United States
- R01 CA013202/CA/NCI NIH HHS/United States
- GM-62580/GM/NIGMS NIH HHS/United States
- CA-13202/CA/NCI NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- R37 CA013202/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous