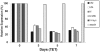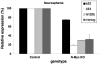N-Myc regulates expression of pluripotency genes in neuroblastoma including lif, klf2, klf4, and lin28b - PubMed (original) (raw)
N-Myc regulates expression of pluripotency genes in neuroblastoma including lif, klf2, klf4, and lin28b
Rebecca Cotterman et al. PLoS One. 2009.
Abstract
myc genes are best known for causing tumors when overexpressed, but recent studies suggest endogenous myc regulates pluripotency and self-renewal of stem cells. For example, N-myc is associated with a number of tumors including neuroblastoma, but also plays a central role in the function of normal neural stem and precursor cells (NSC). Both c- and N-myc also enhance the production of induced pluripotent stem cells (iPSC) and are linked to neural tumor stem cells. The mechanisms by which myc regulates normal and neoplastic stem-related functions remain largely open questions. Here from a global, unbiased search for N-Myc bound genes using ChIP-chip assays in neuroblastoma, we found lif as a putative N-Myc bound gene with a number of strong N-Myc binding peaks in the promoter region enriched for E-boxes. Amongst putative N-Myc target genes in expression microarray studies in neuroblastoma we also found lif and three additional important embryonic stem cell (ESC)-related factors that are linked to production of iPSC: klf2, klf4, and lin28b. To examine the regulation of these genes by N-Myc, we measured their expression using neuroblastoma cells that contain a Tet-regulatable N-myc transgene (TET21N) as well as NSC with a nestin-cre driven N-myc knockout. N-myc levels closely correlated with the expression of all of these genes in neuroblastoma and all but lif in NSC. Direct ChIP assays also indicate that N-Myc directly binds the lif promoter. N-Myc regulates trimethylation of lysine 4 of histone H3 in the promoter of lif and possibly in the promoters of several other stem-related genes. Together these findings indicate that N-Myc regulates overlapping stem-related gene expression programs in neuroblastoma and NSC, supporting a novel model by which amplification of the N-myc gene may drive formation of neuroblastoma. They also suggest mechanisms by which Myc proteins more generally contribute to maintenance of pluripotency and self-renewal of ESC as well as to iPSC formation.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
Figure 1. ChIP-chip indicates strong N-Myc binding of the lif promoter in human neuroblastoma.
ChIP-chip data from previous study was analyzed specifically for lif binding by N-myc. Several strong peaks were observed (brown). The lif promoter is greatly enriched in E-boxes including one canonical myc E-box CACGTG shown in purple and 6 non-canonical indicated by downward vertical arrows and listed in tabular form on the bottom left side. The 3 horizontal black bars labeled “a, b, and c” represent the locations of the 3 PCR products from ChIP PCR reactions in Fig. 4.The genomic locations are listed next to the E-boxes and refer to the last 4 digits of the location, with all being in the 28,970,000 base-pair range on chromosome 22.
Figure 2. Expression of klf2, klf4, lif, and lin28b are linked to N-myc levels in human neuroblastoma.
qRT-PCR quantifies the changes in klf2, klf4, lif, and lin28b expression with similar 3–5 fold decreases in expression for each at each time point of Tet treatment (added daily). Error bars are standard deviation (S.D.) and where not evident it is because the S.D. is so low the bars are so small they do not show up. S.D. throughout this study were quite low and generally ranged from 0.5–5.5%. TET21N neuroblastoma were treated daily with Tet in this experiment. The data here were analyzed using the comparative Ct method, but then reanalyzed using the Pffafl method giving essentially identical results (Fig. S1).
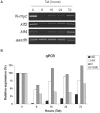
Figure 3. N-myc loss rapidly induces loss of expression of klf2, klf4, lif, and lin28b.
(A) RT-PCR for N-myc, klf2, and klf4 was conducted on Tet21N neuroblastoma cells treated for 8, 16, 24, and 72 hours were conducted along with a loading control (aasdh, a gene we have found does not vary with modulation of myc levels, for example in the TET21N array experiment). N-myc levels recovered from 8 hours onward due to a suboptimal dosing of active Tet. (B) qPCR indicates that klf2, klf4, lif, and lin28b levels rapidly decreased as after only 8 hours of Tet treatment and consequent short-term loss of N-myc. Error bars are S.D.
Figure 4. Chromatin immunoprecipitation (ChIP) assays indicate that N-Myc directly binds lif and klf4 in neuroblastoma.
ChIP was conducted on Tet21N cells treated daily (3 days) or untreated with Tet. IgG was included as a control along with a 1∶50 and 1∶200 dilutions of input. The ChIP'd lif regions a, b, and c are represented by the black bars in Fig. 1. IgG and Input samples were run in parallel as controls.
Figure 5. N-Myc regulates klf2, klf4, and lin28b as well as nanog, but not lif in neurospheres.
Control (N-myc flox/flox) and N-myc null (floxed, nestin-cre) neurospheres were used for qRT-PCR assays. Expression levels in controls were set to 100%. Error bars are S.D.
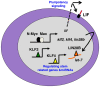
Figure 6. A model of Myc stem-related function in neuroblastoma cells.
Two key functions are depicted: growth factor signaling through LIF and induction of stem-related gene expression through induced transcription of genes encoding KLF2, KLF4, and LIN28B. Together these programs are predicted to maintain a “blast”-like state in neuroblastoma tumors through transcriptional and miRNA functions.
Similar articles
- myc maintains embryonic stem cell pluripotency and self-renewal.
Varlakhanova NV, Cotterman RF, deVries WN, Morgan J, Donahue LR, Murray S, Knowles BB, Knoepfler PS. Varlakhanova NV, et al. Differentiation. 2010 Jul;80(1):9-19. doi: 10.1016/j.diff.2010.05.001. Epub 2010 May 27. Differentiation. 2010. PMID: 20537458 Free PMC article. - Stat3 and c-Myc genome-wide promoter occupancy in embryonic stem cells.
Kidder BL, Yang J, Palmer S. Kidder BL, et al. PLoS One. 2008;3(12):e3932. doi: 10.1371/journal.pone.0003932. Epub 2008 Dec 11. PLoS One. 2008. PMID: 19079543 Free PMC article. - N-myc oncogene overexpression down-regulates leukemia inhibitory factor in neuroblastoma.
Hatzi E, Murphy C, Zoephel A, Ahorn H, Tontsch U, Bamberger AM, Yamauchi-Takihara K, Schweigerer L, Fotsis T. Hatzi E, et al. Eur J Biochem. 2002 Aug;269(15):3732-41. doi: 10.1046/j.1432-1033.2002.03066.x. Eur J Biochem. 2002. PMID: 12153570 - Roles for MYC in the establishment and maintenance of pluripotency.
Chappell J, Dalton S. Chappell J, et al. Cold Spring Harb Perspect Med. 2013 Dec 1;3(12):a014381. doi: 10.1101/cshperspect.a014381. Cold Spring Harb Perspect Med. 2013. PMID: 24296349 Free PMC article. Review. - Myc transcription factors: key regulators behind establishment and maintenance of pluripotency.
Smith K, Dalton S. Smith K, et al. Regen Med. 2010 Nov;5(6):947-59. doi: 10.2217/rme.10.79. Regen Med. 2010. PMID: 21082893 Free PMC article. Review.
Cited by
- A MYCN-driven de-differentiation profile identifies a subgroup of aggressive retinoblastoma.
Ryl T, Afanasyeva E, Hartmann T, Schwermer M, Schneider M, Schröder C, Wagemanns M, Bister A, Kanber D, Steenpass L, Schramm K, Jones B, Jones DTW, Biewald E, Astrahantseff K, Hanenberg H, Rahmann S, Lohmann DR, Schramm A, Ketteler P. Ryl T, et al. Commun Biol. 2024 Jul 30;7(1):919. doi: 10.1038/s42003-024-06596-6. Commun Biol. 2024. PMID: 39079981 Free PMC article. - KR158 Spheres Harboring Slow-Cycling Cells Recapitulate High-Grade Glioma Features in an Immunocompetent System.
Chakraborty A, Yang C, Kresak JL, Silver AJ, Feier D, Tian G, Andrews M, Sobanjo OO, Hodge ED, Engelbart MK, Huang J, Harrison JK, Sarkisian MR, Mitchell DA, Deleyrolle LP. Chakraborty A, et al. Cells. 2024 May 29;13(11):938. doi: 10.3390/cells13110938. Cells. 2024. PMID: 38891070 Free PMC article. - A multi-omics approach for biomarker discovery in neuroblastoma: a network-based framework.
Hussein R, Abou-Shanab AM, Badr E. Hussein R, et al. NPJ Syst Biol Appl. 2024 May 17;10(1):52. doi: 10.1038/s41540-024-00371-3. NPJ Syst Biol Appl. 2024. PMID: 38760476 Free PMC article. - DFMO inhibition of neuroblastoma tumorigenesis.
Gandra D, Mulama DH, Foureau DM, McKinney KQ, Kim E, Smith K, Haw J, Nagulapally A, Saulnier Sholler GL. Gandra D, et al. Cancer Med. 2024 May;13(9):e7207. doi: 10.1002/cam4.7207. Cancer Med. 2024. PMID: 38686627 Free PMC article. - Differentiated neuroblastoma cells remain epigenetically poised for de-differentiation to an immature state.
Guyer RA, Picard N, Mueller JL, Ohishi K, Leavitt A, Murphy AJ, Cornejo KM, Hotta R, Goldstein AM. Guyer RA, et al. Dis Model Mech. 2023 Dec 1;16(12):dmm049754. doi: 10.1242/dmm.049754. Epub 2023 Dec 27. Dis Model Mech. 2023. PMID: 38095019 Free PMC article.
References
- Guccione E, Martinato F, Finocchiaro G, Luzi L, Tizzoni L, et al. Myc-binding-site recognition in the human genome is determined by chromatin context. Nat Cell Biol. 2006;8:764–770. - PubMed
- Cawley S, Bekiranov S, Ng HH, Kapranov P, Gingeras TR. Unbiased mapping of transcription factor binding sites along human chromosomes 21 and 22 points to widespread regulation of noncoding RNAs. Cell. 2004;116:499–509. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical