PubChem: a public information system for analyzing bioactivities of small molecules - PubMed (original) (raw)
. 2009 Jul;37(Web Server issue):W623-33.
doi: 10.1093/nar/gkp456. Epub 2009 Jun 4.
Affiliations
- PMID: 19498078
- PMCID: PMC2703903
- DOI: 10.1093/nar/gkp456
PubChem: a public information system for analyzing bioactivities of small molecules
Yanli Wang et al. Nucleic Acids Res. 2009 Jul.
Abstract
PubChem (http://pubchem.ncbi.nlm.nih.gov) is a public repository for biological properties of small molecules hosted by the US National Institutes of Health (NIH). PubChem BioAssay database currently contains biological test results for more than 700 000 compounds. The goal of PubChem is to make this information easily accessible to biomedical researchers. In this work, we present a set of web servers to facilitate and optimize the utility of biological activity information within PubChem. These web-based services provide tools for rapid data retrieval, integration and comparison of biological screening results, exploratory structure-activity analysis, and target selectivity examination. This article reviews these bioactivity analysis tools and discusses their uses. Most of the tools described in this work can be directly accessed at http://pubchem.ncbi.nlm.nih.gov/assay/. URLs for accessing other tools described in this work are specified individually.
Figures
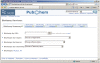
Figure 1.
Common gateway of PubChem BioActivity Analysis Service. It provides a central entry point for accessing bioassay records, and tools including BioAssay Summary, BioActivity Summary, Data Table and Structure–Activity Analysis. Files saved for recording analysis status can be imported using the ‘Open Saved View’ tab.
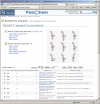
Figure 2.
The BioAssay Summary view for PubChem assay AID 523, a screening experiment aimed to identify inhibitors of human liver cathepsin B, a lysosomal cysteine protease which is associated with several human diseases. PubChem provides similar summary for each of the bioassay records, which can be accessed at the same url shown by this screen shot with the respective AID.
Figure 3.
A view of BioActivity Summary analysis for the 10 compounds active in the secondary assay ‘Cathepsin B Inhibitor Series SAR Study’ (AID 523). This analysis shows that these compounds are tested in several hundreds of screening experiments, and some compounds are considered active in other biological tests as well. A further analysis by revising the assay focus using the ‘Select Active’ tool will tell the number of assays having one or multiple hits overlapping with those in AID 523. To review compounds and the test results from the ‘HTS to identify Inhibitors of West Nile Virus’ assay, AID 577, for example, one may click on ‘6’ shown under the ‘Active’ column to retrieve assay results given in AID 577 for the six compounds which are considered active in that assay. Alternatively, one may select assays using the check box on left of AID, and invoke the ‘Data Table’ tab to retrieve test results from the selected assays.
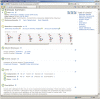
Figure 4.
Screen shot of PubChem structure-activity analysis for the 10 active compounds in AID 523 and several confirmatory assays against a few protein targets. Assay clusters, based on assay target sequence similarity, are shown in the horizontal dimension, while compound clusters, based on 2D structure similarity, are shown in the vertical dimension. Each cell in the heatmap is colored based on the reported active concentration value (e.g. IC50) according to the legend contained within the figure. The PubChem Compound accession, e.g. CID, is shown to the right of each leaf of the compound cluster dendrogram. A chemical structure display can be invoked upon mouse-over the respective CID. PubChem BioAssay accession, e.g. AID, is shown beneath each leaf of the assay cluster dendrogram, while GI numbers for the respective assay targets are provided below the heatmap and hyperlinked to the corresponding Entrez protein records.
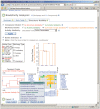
Figure 5.
Concise view of data table for multiple screening test results. The compound centric view merges test results from multiple bioassay depositions, and as a result, one CID may point to multiple SID. Bioactivity outcomes are presented using graphical icons, with ‘active’ results represented with icon colored in red. Complete assay results can be obtained through the ‘Data Table, Complete’ link. One may access additional data analysis tools using the provided links. One may also save the results or analysis status using the features provided in this page (not shown).
Figure 6.
A screen shot of BLAST search results against PubChem BioAssay target database. This report highlights a BLAST hit if biological screening data is available in PubChem for that protein. Each of such BLAST hits is linked to the respective PubChem BioAssay records through a ‘dose–response curve’ icon.
Similar articles
- An overview of the PubChem BioAssay resource.
Wang Y, Bolton E, Dracheva S, Karapetyan K, Shoemaker BA, Suzek TO, Wang J, Xiao J, Zhang J, Bryant SH. Wang Y, et al. Nucleic Acids Res. 2010 Jan;38(Database issue):D255-66. doi: 10.1093/nar/gkp965. Epub 2009 Nov 19. Nucleic Acids Res. 2010. PMID: 19933261 Free PMC article. - PubChem's BioAssay Database.
Wang Y, Xiao J, Suzek TO, Zhang J, Wang J, Zhou Z, Han L, Karapetyan K, Dracheva S, Shoemaker BA, Bolton E, Gindulyte A, Bryant SH. Wang Y, et al. Nucleic Acids Res. 2012 Jan;40(Database issue):D400-12. doi: 10.1093/nar/gkr1132. Epub 2011 Dec 2. Nucleic Acids Res. 2012. PMID: 22140110 Free PMC article. - PubChem Substance and Compound databases.
Kim S, Thiessen PA, Bolton EE, Chen J, Fu G, Gindulyte A, Han L, He J, He S, Shoemaker BA, Wang J, Yu B, Zhang J, Bryant SH. Kim S, et al. Nucleic Acids Res. 2016 Jan 4;44(D1):D1202-13. doi: 10.1093/nar/gkv951. Epub 2015 Sep 22. Nucleic Acids Res. 2016. PMID: 26400175 Free PMC article. - PubChem as a public resource for drug discovery.
Li Q, Cheng T, Wang Y, Bryant SH. Li Q, et al. Drug Discov Today. 2010 Dec;15(23-24):1052-7. doi: 10.1016/j.drudis.2010.10.003. Epub 2010 Oct 21. Drug Discov Today. 2010. PMID: 20970519 Free PMC article. Review. - Education resources of the National Center for Biotechnology Information.
Cooper PS, Lipshultz D, Matten WT, McGinnis SD, Pechous S, Romiti ML, Tao T, Valjavec-Gratian M, Sayers EW. Cooper PS, et al. Brief Bioinform. 2010 Nov;11(6):563-9. doi: 10.1093/bib/bbq022. Epub 2010 Jun 22. Brief Bioinform. 2010. PMID: 20570844 Free PMC article. Review.
Cited by
- Profiling lipid-protein interactions using nonquenched fluorescent liposomal nanovesicles and proteome microarrays.
Lu KY, Tao SC, Yang TC, Ho YH, Lee CH, Lin CC, Juan HF, Huang HC, Yang CY, Chen MS, Lin YY, Lu JY, Zhu H, Chen CS. Lu KY, et al. Mol Cell Proteomics. 2012 Nov;11(11):1177-90. doi: 10.1074/mcp.M112.017426. Epub 2012 Jul 26. Mol Cell Proteomics. 2012. PMID: 22843995 Free PMC article. - Network pharmacology strategies toward multi-target anticancer therapies: from computational models to experimental design principles.
Tang J, Aittokallio T. Tang J, et al. Curr Pharm Des. 2014;20(1):23-36. doi: 10.2174/13816128113199990470. Curr Pharm Des. 2014. PMID: 23530504 Free PMC article. Review. - Data mining of PubChem bioassay records reveals diverse OXPHOS inhibitory chemotypes as potential therapeutic agents against ovarian cancer.
Sharma S, Feng L, Boonpattrawong N, Kapur A, Barroilhet L, Patankar MS, Ericksen SS. Sharma S, et al. J Cheminform. 2024 Oct 7;16(1):112. doi: 10.1186/s13321-024-00906-0. J Cheminform. 2024. PMID: 39375760 Free PMC article. - In silico identification of anti-cancer compounds and plants from traditional Chinese medicine database.
Dai SX, Li WX, Han FF, Guo YC, Zheng JJ, Liu JQ, Wang Q, Gao YD, Li GH, Huang JF. Dai SX, et al. Sci Rep. 2016 May 5;6:25462. doi: 10.1038/srep25462. Sci Rep. 2016. PMID: 27145869 Free PMC article. - In silico approach to screen compounds active against parasitic nematodes of major socio-economic importance.
Khanna V, Ranganathan S. Khanna V, et al. BMC Bioinformatics. 2011;12 Suppl 13(Suppl 13):S25. doi: 10.1186/1471-2105-12-S13-S25. Epub 2011 Nov 30. BMC Bioinformatics. 2011. PMID: 22373185 Free PMC article.
References
- Bolton EE, Wang Y, Thiessen PA, Bryant SH. PubChem: integrated platform of small molecules and biological activities. Annu. Rep. Comput. Chem. 2008;4:217–241. Chapter 12.
- Zerhouni E. Medicine. The NIH roadmap. Science. 2003;302:63–72. - PubMed
- Zerhouni EA. Clinical research at a crossroads: the NIH roadmap. J. Investig. Med. 2006;54:171–173. - PubMed
- Austin CP, Brady LS, Insel TR, Collins FS. NIH molecular libraries initiative. Science. 2004;306:1138–1139. - PubMed
- Lazo JS, Brady LS, Dingledine R. Building a pharmacological lexicon: small molecule discovery in academia. Mol. Pharmacol. 2007;72:1–7. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources