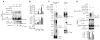Rhes, a striatal specific protein, mediates mutant-huntingtin cytotoxicity - PubMed (original) (raw)
Rhes, a striatal specific protein, mediates mutant-huntingtin cytotoxicity
Srinivasa Subramaniam et al. Science. 2009.
Abstract
Huntington's disease (HD) is caused by a polyglutamine repeat in the protein huntingtin (Htt) with mutant Htt (mHtt) expressed throughout the body and similarly in all brain regions. Yet, HD neuropathology is largely restricted to the corpus striatum. We report that the small guanine nucleotide-binding protein Rhes, which is localized very selectively to the striatum, binds physiologically to mHtt. Using cultured cells, we found Rhes induces sumoylation of mHtt, which leads to cytotoxicity. Thus, Rhes-mHtt interactions can account for the localized neuropathology of HD.
Figures
Fig. 1
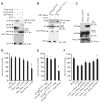
Rhes binds Htt and affects cell survival. (A) Rhes interacts with N-terminal Htt. HEK293 cells were transfected with glutathione S-transferase (GST) or GST-Rhes together with Flag-tagged Htt or the N-terminal fragment containing 171 amino acids and 18 glutamines (wtHtt) or 82 glutamines (mHtt). After 48 hours, cell lysates were glutathione (GSH) precipitated and immunoblotted (IB) for Flag. (B) Rhes interacts with full-length Htt. Striatal cells expressing wild-type Htt (ST_HdhQ7/Q7_) or mutant Htt (ST_HdhQ111/Q111_) were transfected with GST or GST-Rhes. After 48 hours, cell lysates were GSH-precipitated and immunoblotted for Htt. Htt and GST inputs are shown. (C) Rhes interacts with mHtt in striatum. Striatum of transgenic mice expressing mHtt was lysed and immunoprecipitated with Rhes antibody or immunoglobulin IgG alone (bead control). Immunoprecipitates were probed with an N-terminal–specific Htt antibody (N-Htt). (D) Rhes reduces cell survival. HEK293 cells were transfected with Myc/Myc-Rhes and wtHtt–mHtt constructs. ***P < 0.005 versus mHtt alone. (E) Wild-type (ST_HdhQ7/Q7_) or mutant (ST_HdhQ111/Q111_) striatal cells were transfected with Myc/Myc-Rhes. ***P < 0.005 versus Myc. (F) Depletion of Rhes prevents PC12 cell death. Control short hairpin–mediated (shRNA) or Rhes shRNA 1 to 4 were cotransfected with mHtt. Only shRNA4 was significantly cytoprotective (**P < 0.01 versus control shRNA). After 48 hours, cell survival was measured by MTT.
Fig. 2
Rhes inhibits mHtt aggregate formation. (A) HEK293 cells were transfected with Myc or Myc-Rhes and Flag-mHtt. At the indicated time points, cells were lysed, the pellet fraction (containing mHtt aggregates plus SDS-soluble mHtt), and the soluble fractions (containing only soluble Htt) were immunoblotted (IB) for Flag or Rhes. (B) Quantification of aggregated and soluble Htt. Fold change compared with results at 18 hours. (C) Rhes increases mHtt sumoylation in cells. HEK293 cells were transfected with Myc or Myc-Rhes, His-SUMO1 and Flag-mHtt. After 36 hours, cell lysates were immunoprecipitated (IP) with antibody against Flag-IgG beads and immunoblotted for Flag. SUMO-tagged proteins were enriched with TALON metal-affinity resin and immunoblotted for Flag or SUMO1. Input lysates were immunoblotted for SUMO1, Myc, or Flag. (D) Effect of mHtt-K6,9,15,91R mutation on sumoylation and cell survival. HEK293 cells were transfected with GST or GST-Rhes, His-SUMO1, and Myc-tagged mHtt or Myc–mHtt-K6,9,15,91R. After 48 hours, the SUMO-tagged protein was enriched with TALON metal-affinity resin and immunoblotted for Myc. The pellet fraction of the cell lysate was subjected to aggregate detection assay. Cell survival was measured by MTT.
Fig. 3
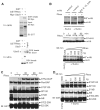
Rhes enhances sumoylation. (A) Rhes interacts with Ubc9. HEK293 cells were transfected with GST or GST-Rhes and Myc-Ubc9. After 48 hours, cells were lysed, precipitated with GSH beads and probed for Myc. Purified GST or GST-Rhes was incubated with purified Ubc9 and precipitated with GSH beads. The precipitates and inputs were immunoblotted for Ubc9. (B) Rhes sumoylates mHtt. Four μl of in vitro translated mHtt (control) and its K6,9,15,91R mutant were subjected to sumoylation [1× RB buffer, 250 ng E1, 125 ng E2, 1.5 μg SUMO1, 5 mM adenosine triphosphate (ATP), and 2 mM dithiothreitol (DTT)] in the presence of 500 ng Rhes (+) or bovine serum albumin (BSA) (−). Htt was detected by N-Htt antibody. Rhes sumoylates Htt in a time- (Rhes, 500 ng) and concentration-dependent manner. (C and D) Sumoylation of multiple substrates. Indicated substrates (500 ng) were subjected to sumoylation assay as in (B). Rhes sumoylates substrates in a (C) time- (Rhes, 200 ng) and (D) concentration-dependent manner.
Fig. 4
Rhes function requires cysteine 263. (A) Disaggregation and cell death. GST or GST-tagged Rhes (WT, C263S, or S33N) were transfected along with Flag-mHtt; Htt aggregation and cell survival were assessed at 48 hours. ***P < 0.001 versus WT. (B) Sumoylation. HEK293 cells were transfected with GST, GST-tagged Rhes (WT, C263S, or S33N), His-SUMO1, and mHtt. After 36 hours, cell lysates were either immunoprecipitated (IP) with Flag-IgG beads or enriched with TALON metal-affinity resin. The precipitate and input were immunoblotted (IB) for Flag, SUMO1, or GST. (C) Rhes/Ubc9/mHtt colocalization. ST_HdhQ111/Q111_ cells were transfected with GST-Rhes WT, C263S, or S33N mutants. After 48 hours, cells were processed for nuclear staining and immunostaining with antibodies against Rhes, Ubc9, and Htt. (D) Rhes/Ubc9/mHtt interaction. ST_HdhQ111/Q111_ cells were transfected with GST or GST-Rhes with WT, S33N, or C263S. After 48 hours, cells were lysed and precipitated with GSH beads. The precipitates and inputs were probed for mHtt, Ubc9, or GST.
Similar articles
- Bioinformatics analysis of Ras homologue enriched in the striatum, a potential target for Huntington's disease therapy.
Carbo M, Brandi V, Pascarella G, Staid DS, Colotti G, Polticelli F, Ilari A, Morea V. Carbo M, et al. Int J Mol Med. 2019 Dec;44(6):2223-2233. doi: 10.3892/ijmm.2019.4373. Epub 2019 Oct 15. Int J Mol Med. 2019. PMID: 31638189 Free PMC article. - The role of Rhes, Ras homolog enriched in striatum, in neurodegenerative processes.
Harrison LM, Lahoste GJ. Harrison LM, et al. Exp Cell Res. 2013 Sep 10;319(15):2310-5. doi: 10.1016/j.yexcr.2013.03.033. Epub 2013 Apr 10. Exp Cell Res. 2013. PMID: 23583659 Review. - Huntington's disease is a disorder of the corpus striatum: focus on Rhes (Ras homologue enriched in the striatum).
Subramaniam S, Snyder SH. Subramaniam S, et al. Neuropharmacology. 2011 Jun;60(7-8):1187-92. doi: 10.1016/j.neuropharm.2010.10.025. Epub 2010 Oct 31. Neuropharmacology. 2011. PMID: 21044641 Review. - Ectopic expression of the striatal-enriched GTPase Rhes elicits cerebellar degeneration and an ataxia phenotype in Huntington's disease.
Swarnkar S, Chen Y, Pryor WM, Shahani N, Page DT, Subramaniam S. Swarnkar S, et al. Neurobiol Dis. 2015 Oct;82:66-77. doi: 10.1016/j.nbd.2015.05.011. Epub 2015 Jun 3. Neurobiol Dis. 2015. PMID: 26048156 - SUMO modification of Huntingtin and Huntington's disease pathology.
Steffan JS, Agrawal N, Pallos J, Rockabrand E, Trotman LC, Slepko N, Illes K, Lukacsovich T, Zhu YZ, Cattaneo E, Pandolfi PP, Thompson LM, Marsh JL. Steffan JS, et al. Science. 2004 Apr 2;304(5667):100-4. doi: 10.1126/science.1092194. Science. 2004. PMID: 15064418
Cited by
- Ubiquitin-dependent and independent roles of SUMO in proteostasis.
Liebelt F, Vertegaal AC. Liebelt F, et al. Am J Physiol Cell Physiol. 2016 Aug 1;311(2):C284-96. doi: 10.1152/ajpcell.00091.2016. Epub 2016 Jun 22. Am J Physiol Cell Physiol. 2016. PMID: 27335169 Free PMC article. Review. - The emerging role of the first 17 amino acids of huntingtin in Huntington's disease.
Arndt JR, Chaibva M, Legleiter J. Arndt JR, et al. Biomol Concepts. 2015 Mar;6(1):33-46. doi: 10.1515/bmc-2015-0001. Biomol Concepts. 2015. PMID: 25741791 Free PMC article. Review. - Bioinformatics analysis of Ras homologue enriched in the striatum, a potential target for Huntington's disease therapy.
Carbo M, Brandi V, Pascarella G, Staid DS, Colotti G, Polticelli F, Ilari A, Morea V. Carbo M, et al. Int J Mol Med. 2019 Dec;44(6):2223-2233. doi: 10.3892/ijmm.2019.4373. Epub 2019 Oct 15. Int J Mol Med. 2019. PMID: 31638189 Free PMC article. - Allosteric Inhibition of Ubiquitin-like Modifications by a Class of Inhibitor of SUMO-Activating Enzyme.
Li YJ, Du L, Wang J, Vega R, Lee TD, Miao Y, Aldana-Masangkay G, Samuels ER, Li B, Ouyang SX, Colayco SA, Bobkova EV, Divlianska DB, Sergienko E, Chung TDY, Fakih M, Chen Y. Li YJ, et al. Cell Chem Biol. 2019 Feb 21;26(2):278-288.e6. doi: 10.1016/j.chembiol.2018.10.026. Epub 2018 Dec 20. Cell Chem Biol. 2019. PMID: 30581133 Free PMC article. - Deciphering the SUMO code in the kidney.
Yang Z, Zhang Y, Sun S. Yang Z, et al. J Cell Mol Med. 2019 Feb;23(2):711-719. doi: 10.1111/jcmm.14021. Epub 2018 Dec 1. J Cell Mol Med. 2019. PMID: 30506859 Free PMC article. Review.
References
- Gusella JF, Macdonald ME. Trends Biochem Sci. 2006;31:533. - PubMed
- Cowan CM, Raymond LA. Curr Top Dev Biol. 2006;75:25. - PubMed
- Sieradzan KA, Mann DM. Neuropathol Appl Neurobiol. 2001;27:1. - PubMed
- Saudou F, Finkbeiner S, Devys D, Greenberg ME. Cell. 1998;95:55. - PubMed
- Kuemmerle S, et al. Ann Neurol. 1999;46:842. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- DA00074/DA/NIDA NIH HHS/United States
- R37 MH018501/MH/NIMH NIH HHS/United States
- K05 DA000074/DA/NIDA NIH HHS/United States
- MH18501/MH/NIMH NIH HHS/United States
- R01 MH018501/MH/NIMH NIH HHS/United States
- R37 MH018501-40/MH/NIMH NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials