AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading - PubMed (original) (raw)
AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading
Oleg Trott et al. J Comput Chem. 2010.
Abstract
AutoDock Vina, a new program for molecular docking and virtual screening, is presented. AutoDock Vina achieves an approximately two orders of magnitude speed-up compared with the molecular docking software previously developed in our lab (AutoDock 4), while also significantly improving the accuracy of the binding mode predictions, judging by our tests on the training set used in AutoDock 4 development. Further speed-up is achieved from parallelism, by using multithreading on multicore machines. AutoDock Vina automatically calculates the grid maps and clusters the results in a way transparent to the user.
Copyright 2009 Wiley Periodicals, Inc.
Figures
Figure 1
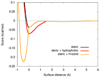
Weighted scoring function term combinations. Steric interactions; steric and hydrophobic interactions; and steric and hydrogen bonding interactions, using weights from table 1
Figure 2
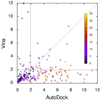
RMSD of the predicted bound ligand conformation from the experimental one, showing the values for Vina and Autodock. The color encodes the number of active rotatable bonds
Figure 3
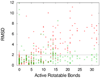
RMSD of the predicted bound ligand conformation from the experimental one. (Red +) Autodock; (Green ×) Vina. Abscissa shows the number of active rotatable bonds
Figure 4
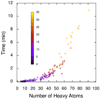
Time, in minutes, taken by the multi-threaded Vina run, as a function of the number of heavy atoms in the ligand. The color encodes the number of active rotatable bonds
Figure 5
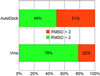
The fraction of the 190 test complexes for which RMSD < 2Å was achieved by AutoDock and Vina
Figure 6
Average time, in minutes per complex, taken by AutoDock, single-threaded Vina and Vina with 8-way multithreading. Machines with two quad-core 2.66GHz Xeon processors were used in the experiment. The time for AutoDock does not include the necessary initial AutoGrid run
Figure 7
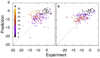
Experimental and predicted free energies of binding (kcal/mol) for (a) AutoDock, (b) Vina. The color encodes the number of active rotatable bonds
Similar articles
- QuickVina: accelerating AutoDock Vina using gradient-based heuristics for global optimization.
Handoko SD, Ouyang X, Su CT, Kwoh CK, Ong YS. Handoko SD, et al. IEEE/ACM Trans Comput Biol Bioinform. 2012 Sep-Oct;9(5):1266-72. doi: 10.1109/TCBB.2012.82. IEEE/ACM Trans Comput Biol Bioinform. 2012. PMID: 22641710 - The Performance of Several Docking Programs at Reproducing Protein-Macrolide-Like Crystal Structures.
Castro-Alvarez A, Costa AM, Vilarrasa J. Castro-Alvarez A, et al. Molecules. 2017 Jan 17;22(1):136. doi: 10.3390/molecules22010136. Molecules. 2017. PMID: 28106755 Free PMC article. - The scoring bias in reverse docking and the score normalization strategy to improve success rate of target fishing.
Luo Q, Zhao L, Hu J, Jin H, Liu Z, Zhang L. Luo Q, et al. PLoS One. 2017 Feb 14;12(2):e0171433. doi: 10.1371/journal.pone.0171433. eCollection 2017. PLoS One. 2017. PMID: 28196116 Free PMC article. - Vinardo: A Scoring Function Based on Autodock Vina Improves Scoring, Docking, and Virtual Screening.
Quiroga R, Villarreal MA. Quiroga R, et al. PLoS One. 2016 May 12;11(5):e0155183. doi: 10.1371/journal.pone.0155183. eCollection 2016. PLoS One. 2016. PMID: 27171006 Free PMC article. - Survey of public domain software for docking simulations and virtual screening.
Biesiada J, Porollo A, Velayutham P, Kouril M, Meller J. Biesiada J, et al. Hum Genomics. 2011 Jul;5(5):497-505. doi: 10.1186/1479-7364-5-5-497. Hum Genomics. 2011. PMID: 21807604 Free PMC article. Review.
Cited by
- Influence of budesonide and fluticasone propionate in the anti-osteoporotic potential in human bone marrow-derived mesenchymal stem cells via stimulation of osteogenic differentiation.
Sumague TS, Niazy AA, Lambarte RNA, Nafisah IA, Gusnanto A. Sumague TS, et al. Heliyon. 2024 Oct 18;10(20):e39475. doi: 10.1016/j.heliyon.2024.e39475. eCollection 2024 Oct 30. Heliyon. 2024. PMID: 39497989 Free PMC article. - An in-silico approach to target multiple proteins involved in anti-microbial resistance using natural compounds produced by wild mushrooms.
Singh G, Hossain MA, Al-Fahad D, Gupta V, Tandon S, Soni H, Narasimhaji CV, Jaremko M, Emwas AH, Anwar MJ, Azam F. Singh G, et al. Biochem Biophys Rep. 2024 Oct 21;40:101854. doi: 10.1016/j.bbrep.2024.101854. eCollection 2024 Dec. Biochem Biophys Rep. 2024. PMID: 39498442 Free PMC article. - Glucosinolates and Indole-3-carbinol from Brassica oleracea L. as inhibitors of E. coli CdtB: insights from molecular docking, dynamics, DFT and in vitro assay.
Tasnim F, Hosen ME, Haque ME, Islam A, Nuryay MN, Mawya J, Akter N, Yesmin D, Hossain MM, Rahman N, Mahmudul Hasan BM, Hassan MN, Islam MM, Khalekuzzaman M. Tasnim F, et al. In Silico Pharmacol. 2024 Oct 29;12(2):95. doi: 10.1007/s40203-024-00276-3. eCollection 2024. In Silico Pharmacol. 2024. PMID: 39479380 - Siteng Fang Reverses Multidrug Resistance in Gastric Cancer: A Network Pharmacology and Molecular Docking Study.
Guo L, Shi H, Zhu L. Guo L, et al. Front Oncol. 2021 May 7;11:671382. doi: 10.3389/fonc.2021.671382. eCollection 2021. Front Oncol. 2021. PMID: 34026648 Free PMC article. - Enhanced Accumulation of Cisplatin in Ovarian Cancer Cells from Combination with Wedelolactone and Resulting Inhibition of Multiple Epigenetic Drivers.
Sarwar S, Alamro AA, Alghamdi AA, Naeem K, Ullah S, Arif M, Yu JQ, Huq F. Sarwar S, et al. Drug Des Devel Ther. 2021 May 25;15:2211-2227. doi: 10.2147/DDDT.S288707. eCollection 2021. Drug Des Devel Ther. 2021. PMID: 34079223 Free PMC article.
References
- Sousa SF, Fernandes PA, Ramos MJ. Protein-ligand docking: Current status and future challenges. Proteins-Structure Function and Bioinformatics. 2006 Oct 1;vol. 65:15–26. - PubMed
- Chang MW, Lindstrom W, Olson AJ, Belew RK. Analysis of HIV wild-type and mutant structures via in silico docking against diverse ligand libraries. Journal of Chemical Information and Modeling. 2007 May–Jun;vol. 47:1258–1262. - PubMed
- Gilson MK, Zhou H-X. Calculation of protein-ligand binding affinities. Annual Review of Biophysics and Biomolecular Structure. 2007;vol. 36:21–42. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous