Solution characterization of the extracellular region of CD147 and its interaction with its enzyme ligand cyclophilin A - PubMed (original) (raw)
. 2009 Aug 21;391(3):518-35.
doi: 10.1016/j.jmb.2009.05.080. Epub 2009 Jun 3.
Jasmina S Redzic, Christopher C Porter, Vyacheslav Yurchenko, Michael Bukrinsky, Wladimir Labeikovsky, Geoffrey S Armstrong, Fengli Zhang, Nancy G Isern, James DeGregori, Robert Hodges, Elan Zohar Eisenmesser
Affiliations
- PMID: 19500591
- PMCID: PMC2940942
- DOI: 10.1016/j.jmb.2009.05.080
Solution characterization of the extracellular region of CD147 and its interaction with its enzyme ligand cyclophilin A
Jennifer Schlegel et al. J Mol Biol. 2009.
Abstract
The CD147 receptor plays an integral role in numerous diseases by stimulating the expression of several protein families and serving as the receptor for extracellular cyclophilins; however, neither CD147 nor its interactions with its cyclophilin ligands have been well characterized in solution. CD147 is a unique protein in that it can function both at the cell membrane and after being released from cells where it continues to retain activity. Thus, the CD147 receptor functions through at least two mechanisms that include both cyclophilin-independent and cyclophilin-dependent modes of action. In regard to CD147 cyclophilin-independent activity, CD147 homophilic interactions are thought to underlie its activity. In regard to CD147 cyclophilin-dependent activity, cyclophilin/CD147 interactions may represent a novel means of signaling since cyclophilins are also peptidyl-prolyl isomerases. However, direct evidence of catalysis has not been shown within the cyclophilin/CD147 complex. In this report, we have characterized the solution behavior of the two most prevalent CD147 extracellular isoforms through biochemical methods that include gel-filtration and native gel analysis as well as directly through multiple NMR methods. All methods indicate that the extracellular immunoglobulin-like domains are monomeric in solution and, thus, suggest that CD147 homophilic interactions in vivo are mediated through other partners. Additionally, using multiple NMR techniques, we have identified and characterized the cyclophilin target site on CD147 and have shown for the first time that CD147 is also a substrate of its primary cyclophilin enzyme ligand, cyclophilin A.
Figures
Fig. 1
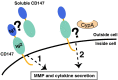
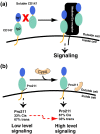
CD147 functions through both cyclophilin-independent and cyclophilin-dependent mechanisms. Both mechanisms stimulate the secretion of MMPs and pro-inflammatory cytokines. These mechanisms include the following: Mechanism 1, CD147 homophilic interactions occur both between neighboring cells as well as soluble CD147 released by cells through several known mechanisms (see the text). Mechanism 2, extracellular cyclophilin enzymes target the CD147 receptor. CD147 Ig-like domains, Ig1 (blue) and Ig2 (green), are shown along with the predicted flexible regions (orange) and CypA (khaki).
Fig. 2
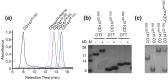
Purified CD147 constructs exist as single monomeric species in solution in the absence of the TM region. (a) Size-exclusion chromatography of a subset of the constructs analyzed in this study and their retention volume upon loading 100 μl of 1 mg/ml of each sample onto an analytical Superose-12 column ( column volume, 24 ml). Only constructs that contain the full TM domain, such as the full-length protein CD14722–269, induce the formation of large oligomers, as shown by their elution at the void volume (∼ 8 ml). The estimated molecular weights of CD14794–205 and CD14722–103 elute near the predicted molecular weights of a monomer, while both CD14722–269Δ™ and CD14722–205 elute higher due to their flexible regions and the flexible nature of the linker region between the two Ig-like domains. (b) Denaturing gel of CD147 constructs containing both domains (CD14722–205), Ig1 (CD14722–103), and Ig2 (CD14794–205) in the absence and presence of DTT. Reduction of the single disulfide bond within each domain leads to slower migratory behavior. (c) Native gel of the same constructs confirms that all constructs run as a single species and that Ig1 and Ig2 do not interact (right lane).
Fig. 3
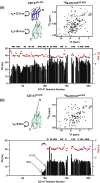
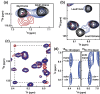
NMR solution studies reveal that the CD147 Ig-like domains do not self-associate or interact with each other. The extracellular regions of the two major CD147 isoforms, (a) isoform 2 (CD14722–205) and (b) isoform 3 (CD14794–205), were characterized using NMR solution studies. The particular regions under study are shown using the X-ray crystal structure (Protein Data Bank accession code: 3B5H) with the corresponding 15N-HSQC spectra and NMR relaxation data. Specifically, amide R1 (red dots) and R2 (black bars) relaxation rates were obtained for both CD147 constructs and used to calculate the correlation times. The extracted correlation times are shown adjacent to each Ig-like domain, Ig1 (blue) and Ig2 (green). Secondary-structure propensities are shown on top of each plot as calculated from our chemical shift assignments with the program TALOS. All data were collected at 25 °C and 900 MHz.
Fig. 4
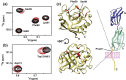
CypA specifically engages CD147 through its active site. (a) 15N-TROSY-HSQC spectra of 0.5 mM free 15N-CypA (black) and in the presence of 1 mM CD14722–214 (red) indicate that only the chemical environment of CypA active-site residues such as Ser99 and Phe60 are perturbed by the interaction. (b) Within these same spectra, 15N-CypA Trp121Nɛ1 that is located in the short 3,10–α-helix adjacent to the active site also exhibits chemical shift perturbation in the presence of CD14722–214. (c) Two views of the CypA residues that exhibit chemical shift perturbations upon addition of CD14722–214 are shown, along with a cartoon extension of the Ig-like domains that contain the targeted Pro211. Residues are colored (red) onto our previously determined X-ray crystal structure of CypA (Protein Data Bank accession code: 1ZKT) if their respective chemical shift differences were greater than 0.06 ppm as defined by (5Δν1H)2+(Δν15N)2. Titrations were conducted at 25 °C and at 900 MHz.
Fig. 5
Quantifying the binding interaction between CypA and CD147. (a) Binding isotherms are shown for both 15N-CypA active-site amides and the single indole of the active-site Trp121 upon addition of a peptide that corresponds to CD147 residues 205–214, the CD14710mer. A _K_d of 4.2 ± 0.2 mM was extracted from these fits. (b) Chemical shift changes of 15N-CypA are compared upon titration of CD14722–214 (1 mM, red) and the CD14710mer peptide (8 mM, black). (c) CypA chemical shift changes with the CD14710mer peptide are relegated to the active site as they are with the larger CD147 constructs. All chemical shift changes were calculated as in Fig. 4. Experiments were collected at 25 °C and at 900 MHz.
Fig. 6
CD147 is a substrate of CypA. (a) 15N-TROSY-HSQC spectra of 0.5 mM free 15N-CD14794–214 (black) and in the presence of both 100 μM CypA (blue) and 1 mM CypA (red) show that upon titration of the enzyme, CD147 Gly214 amide cis and trans resonances converge. Thus, relative to the CypA-bound chemical shift difference between the CD147 Gly214 cis and trans conformations, the CypA induced conformational exchange rate is fast. (b) Several of the CypA-targeted CD147 residues, such as CD147 Leu213, exhibit severe linebroadening in the presence of substoichiometric concentrations of the enzyme and disappear under stoichiometric concentrations. Thus, relative to the CypA-bound chemical shift differences, the CypA-induced conformational exchange is on the intermediate timescale. (c) Using 0.5 mM 15N-CD14794–214, ZZ-exchange cross peaks are observed for residues 207–214 in the presence of catalytic concentrations of CypA. Shown here is the corresponding spectrum of the CD147 Phe212 amide with 0.05 mM CypA and a mixing time of 0 ms (dark blue) and 300 ms (red). (d) 3D-NOESY-HSQC of 0.5 mM 15N-CD14794–214 and 0.01 mM CypA utilizing a 150-ms mixing time also identifies exchange peaks for CD147 residues 207–214 as shown for Phe212. In the absence of CypA, no such cross peaks are observed for either ZZ-exchange or within 3D-NOESY-HSQC experiments, indicating a much slower uncatalyzed exchange between CD147 cis and trans conformations (data not shown). All experiments were conducted at 25 °C. Titrations were acquired at 900 MHz, and both ZZ-exchange and 3D-NOESY-experiments were acquired at 720 MHz.
Fig. 7
Quantifying the catalytic rate of exchange for CypA-mediated isomerization of CD147. The apparent catalytic exchange rates in Table 1 were derived from ZZ-exchange experiments using [CD14794–214] = 0.5 mM and either (a) [CypA] = 0.01 mM or (b) [CypA] = 0.05 mM. The ZZ-exchange data along with the corresponding fits and two sample spectra from each experiment for CD147 Trp210Nɛ1 are shown. Intensities for each of the four resonances were fit simultaneously to Eqs. (1a), (1b), (1c), and (1d). Intensities were normalized to that of the highest intensity, that is, the trans resonance, at no mixing time.
Fig. 8
Models for both the cyclophilin-independent and cyclophilin-dependent mechanisms of CD147 activity. (a) Based on our findings here that have shown that the extracellular region of CD147 does not self-associate, CD147 homophilic interactions are likely mediated by other molecules that potentially include TM proteins (i.e., co-receptors). (b) The direct observation of CypA-mediated isomerization of Pro211 suggests a mechanism whereby CypA may alter the inherent cis:trans equilibrium of CD147 Pro211. The inherent cis:trans ratio of CD147 Pro211 was calculated from the relative peak intensities of the amides of residues 207–214 within 15N-HSQC spectra, and the cis:trans ratio in the active complex is based on previous studies with a model peptide substrate. Colors are as defined in Fig. 1.
Fig. S2
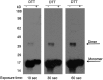
Western blot analysis reveals trace amounts of disulfide-linked CD147. Although not visible in standard SDS-PAGE analysis, Western blot analysis shown here reveals that a small fraction of CD14722–205 refolded from bacterial intracellular expression via our methods exists as a dimer and that this is lost upon addition of reducing agent.
Similar articles
- Cyclophilin-CD147 interactions: a new target for anti-inflammatory therapeutics.
Yurchenko V, Constant S, Eisenmesser E, Bukrinsky M. Yurchenko V, et al. Clin Exp Immunol. 2010 Jun;160(3):305-17. doi: 10.1111/j.1365-2249.2010.04115.x. Epub 2010 Mar 16. Clin Exp Immunol. 2010. PMID: 20345978 Free PMC article. Review. - Cyclophilin A (CyPA) induces chemotaxis independent of its peptidylprolyl cis-trans isomerase activity: direct binding between CyPA and the ectodomain of CD147.
Song F, Zhang X, Ren XB, Zhu P, Xu J, Wang L, Li YF, Zhong N, Ru Q, Zhang DW, Jiang JL, Xia B, Chen ZN. Song F, et al. J Biol Chem. 2011 Mar 11;286(10):8197-8203. doi: 10.1074/jbc.C110.181347. Epub 2011 Jan 18. J Biol Chem. 2011. PMID: 21245143 Free PMC article. - Active site residues of cyclophilin A are crucial for its signaling activity via CD147.
Yurchenko V, Zybarth G, O'Connor M, Dai WW, Franchin G, Hao T, Guo H, Hung HC, Toole B, Gallay P, Sherry B, Bukrinsky M. Yurchenko V, et al. J Biol Chem. 2002 Jun 21;277(25):22959-65. doi: 10.1074/jbc.M201593200. Epub 2002 Apr 9. J Biol Chem. 2002. PMID: 11943775 - Dealing with the family: CD147 interactions with cyclophilins.
Yurchenko V, Constant S, Bukrinsky M. Yurchenko V, et al. Immunology. 2006 Mar;117(3):301-9. doi: 10.1111/j.1365-2567.2005.02316.x. Immunology. 2006. PMID: 16476049 Free PMC article. Review. - Inhibition of the cyclophilin A-CD147 interaction attenuates right ventricular injury and dysfunction after acute pulmonary embolism in rats.
Lu G, Jia Z, Zu Q, Zhang J, Zhao L, Shi H. Lu G, et al. J Biol Chem. 2018 Aug 3;293(31):12199-12208. doi: 10.1074/jbc.RA118.002845. Epub 2018 Jun 18. J Biol Chem. 2018. PMID: 29914983 Free PMC article.
Cited by
- Structural and biochemical characterization of the human cyclophilin family of peptidyl-prolyl isomerases.
Davis TL, Walker JR, Campagna-Slater V, Finerty PJ, Paramanathan R, Bernstein G, MacKenzie F, Tempel W, Ouyang H, Lee WH, Eisenmesser EZ, Dhe-Paganon S. Davis TL, et al. PLoS Biol. 2010 Jul 27;8(7):e1000439. doi: 10.1371/journal.pbio.1000439. PLoS Biol. 2010. PMID: 20676357 Free PMC article. - 1H, 13C, and 15N backbone and side chain resonance assignments of thermophilic Geobacillus kaustophilus cyclophilin-A.
Holliday MJ, Zhang F, Isern NG, Armstrong GS, Eisenmesser EZ. Holliday MJ, et al. Biomol NMR Assign. 2014 Apr;8(1):23-7. doi: 10.1007/s12104-012-9445-3. Epub 2012 Nov 9. Biomol NMR Assign. 2014. PMID: 23138858 Free PMC article. - Interleukin-37 monomer is the active form for reducing innate immunity.
Eisenmesser EZ, Gottschlich A, Redzic JS, Paukovich N, Nix JC, Azam T, Zhang L, Zhao R, Kieft JS, The E, Meng X, Dinarello CA. Eisenmesser EZ, et al. Proc Natl Acad Sci U S A. 2019 Mar 19;116(12):5514-5522. doi: 10.1073/pnas.1819672116. Epub 2019 Feb 28. Proc Natl Acad Sci U S A. 2019. PMID: 30819901 Free PMC article. - Basigin is a receptor essential for erythrocyte invasion by Plasmodium falciparum.
Crosnier C, Bustamante LY, Bartholdson SJ, Bei AK, Theron M, Uchikawa M, Mboup S, Ndir O, Kwiatkowski DP, Duraisingh MT, Rayner JC, Wright GJ. Crosnier C, et al. Nature. 2011 Nov 9;480(7378):534-7. doi: 10.1038/nature10606. Nature. 2011. PMID: 22080952 Free PMC article. - A novel CD147 inhibitor, SP-8356, reduces neointimal hyperplasia and arterial stiffness in a rat model of partial carotid artery ligation.
Pahk K, Noh H, Joung C, Jang M, Song HY, Kim KW, Han K, Hwang JI, Kim S, Kim WK. Pahk K, et al. J Transl Med. 2019 Aug 20;17(1):274. doi: 10.1186/s12967-019-2024-y. J Transl Med. 2019. PMID: 31429778 Free PMC article.
References
- Guo H.M., Majmudar G., Jensen T.C., Biswas C., Toole B.P., Gordon M.K. Characterization of the gene for human EMMPRIN, a tumor cell surface inducer of matrix metalloproteinases. Gene. 1998;220:99–108. - PubMed
- Schmidt R., Bültmann A., Fischel S., Gillitzer A., Cullen P., Walch A. Extracellular matrix metalloproteinase inducer (CD147) is a novel receptor on platelets, activates platelets, and augments nuclear factor {kappa}B dependent inflammation in monocytes. Circ. Res. 2008:107. - PubMed
- Ruiz S., Castro-Castro A., Bustelo X.R. CD147 inhibits the nuclear factor of activated T-cells by impairing vav1 and rac1 downstream signaling. J. Biol. Chem. 2008;283:5554–5566. - PubMed
- Gabison E.E., Hoang-Xuan T., Mauviel A., Menashi S. EMMPRIN/CD147, an MMP modulator in cancer, development and tissue repair. Biochimie. 2005;87:361–368. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R56 AI081152-01A1/AI/NIAID NIH HHS/United States
- R01 CA109657/CA/NCI NIH HHS/United States
- T32 CA082086/CA/NCI NIH HHS/United States
- R56 AI081152/AI/NIAID NIH HHS/United States
- R21 AI060720-01/AI/NIAID NIH HHS/United States
- R01 AI081152-01A1/AI/NIAID NIH HHS/United States
- R21 AI060720/AI/NIAID NIH HHS/United States
- R21 AI060720-02/AI/NIAID NIH HHS/United States
- GM68928/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous