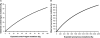Correcting for purifying selection: an improved human mitochondrial molecular clock - PubMed (original) (raw)
Correcting for purifying selection: an improved human mitochondrial molecular clock
Pedro Soares et al. Am J Hum Genet. 2009 Jun.
Abstract
There is currently no calibration available for the whole human mtDNA genome, incorporating both coding and control regions. Furthermore, as several authors have pointed out recently, linear molecular clocks that incorporate selectable characters are in any case problematic. We here confirm a modest effect of purifying selection on the mtDNA coding region and propose an improved molecular clock for dating human mtDNA, based on a worldwide phylogeny of > 2000 complete mtDNA genomes and calibrating against recent evidence for the divergence time of humans and chimpanzees. We focus on a time-dependent mutation rate based on the entire mtDNA genome and supported by a neutral clock based on synonymous mutations alone. We show that the corrected rate is further corroborated by archaeological dating for the settlement of the Canary Islands and Remote Oceania and also, given certain phylogeographic assumptions, by the timing of the first modern human settlement of Europe and resettlement after the Last Glacial Maximum. The corrected rate yields an age of modern human expansion in the Americas at approximately 15 kya that-unlike the uncorrected clock-matches the archaeological evidence, but continues to indicate an out-of-Africa dispersal at around 55-70 kya, 5-20 ky before any clear archaeological record, suggesting the need for archaeological research efforts focusing on this time window. We also present improved rates for the mtDNA control region, and the first comprehensive estimates of positional mutation rates for human mtDNA, which are essential for defining mutation models in phylogenetic analyses.
Figures
Figure 1
Hot Spots in the mtDNA Genome, Showing All the Positions that Appear > 25 Times in the Tree
Figure 2
mtDNA Saturation Curves (A) Control-region mutations. (B) Synonymous mutations.
Figure 3
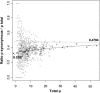
Relation between Overall ρ Values and the Synonymous ρ Proportion Individual data points for each node in the tree are displayed as filled gray circles, and grouped points are displayed as empty circles. The solid curve is a model fit to the empty circles.
Figure 4
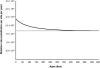
Variation of the mtDNA Mutation Rate through Time The black curve represents the mtDNA mutation rate. The gray curve represents the interspecific mutation rate.
Figure 5
Comparison of the Age of Each Node with the Time-Dependent Complete mtDNA Genome Sequence Molecular Clock and the Synonymous Clock
Figure 6
A New Chronology for the Human mtDNA Tree The tree was obtained by combining six trees constructed separately, with branch lengths estimated with maximum likelihood and the time-dependent molecular clock. Number and presence of clades is dependent on availability of data and not on worldwide frequencies. Ages are expressed in kya.
Similar articles
- A mitogenomic phylogeny and genetic history of sable (Martes zibellina).
Malyarchuk B, Derenko M, Denisova G. Malyarchuk B, et al. Gene. 2014 Oct 15;550(1):56-67. doi: 10.1016/j.gene.2014.08.015. Epub 2014 Aug 7. Gene. 2014. PMID: 25110108 - Population expansion in the North African late Pleistocene signalled by mitochondrial DNA haplogroup U6.
Pereira L, Silva NM, Franco-Duarte R, Fernandes V, Pereira JB, Costa MD, Martins H, Soares P, Behar DM, Richards MB, Macaulay V. Pereira L, et al. BMC Evol Biol. 2010 Dec 21;10:390. doi: 10.1186/1471-2148-10-390. BMC Evol Biol. 2010. PMID: 21176127 Free PMC article. - Explaining the imperfection of the molecular clock of hominid mitochondria.
Loogväli EL, Kivisild T, Margus T, Villems R. Loogväli EL, et al. PLoS One. 2009 Dec 29;4(12):e8260. doi: 10.1371/journal.pone.0008260. PLoS One. 2009. PMID: 20041137 Free PMC article. - Harvesting the fruit of the human mtDNA tree.
Torroni A, Achilli A, Macaulay V, Richards M, Bandelt HJ. Torroni A, et al. Trends Genet. 2006 Jun;22(6):339-45. doi: 10.1016/j.tig.2006.04.001. Epub 2006 May 4. Trends Genet. 2006. PMID: 16678300 Review. - Catarrhine primate divergence dates estimated from complete mitochondrial genomes: concordance with fossil and nuclear DNA evidence.
Raaum RL, Sterner KN, Noviello CM, Stewart CB, Disotell TR. Raaum RL, et al. J Hum Evol. 2005 Mar;48(3):237-57. doi: 10.1016/j.jhevol.2004.11.007. Epub 2005 Jan 20. J Hum Evol. 2005. PMID: 15737392 Review.
Cited by
- Mitochondrial DNA Genomes Reveal Relaxed Purifying Selection During Human Population Expansion after the Last Glacial Maximum.
Zheng HX, Yan S, Zhang M, Gu Z, Wang J, Jin L. Zheng HX, et al. Mol Biol Evol. 2024 Sep 4;41(9):msae175. doi: 10.1093/molbev/msae175. Mol Biol Evol. 2024. PMID: 39162340 Free PMC article. - New Canary Islands Roman mediated settlement hypothesis deduced from coalescence ages of curated maternal indigenous lineages.
Cabrera VM. Cabrera VM. Sci Rep. 2024 May 15;14(1):11150. doi: 10.1038/s41598-024-61731-x. Sci Rep. 2024. PMID: 38750053 Free PMC article. - Extracellular vesicle mitochondrial DNA levels are associated with race and mitochondrial DNA haplogroup.
Byappanahalli AM, Omoniyi V, Noren Hooten N, Smith JT, Mode NA, Ezike N, Zonderman AB, Evans MK. Byappanahalli AM, et al. iScience. 2023 Dec 13;27(1):108724. doi: 10.1016/j.isci.2023.108724. eCollection 2024 Jan 19. iScience. 2023. PMID: 38226163 Free PMC article. - Mitochondrial point heteroplasmy: insights from deep-sequencing of human replicate samples.
Korolija M, Sukser V, Vlahoviček K. Korolija M, et al. BMC Genomics. 2024 Jan 10;25(1):48. doi: 10.1186/s12864-024-09963-z. BMC Genomics. 2024. PMID: 38200446 Free PMC article. - Dating ancient splits in phylogenetic trees, with application to the human-Neanderthal split.
Levinstein Hallak K, Rosset S. Levinstein Hallak K, et al. BMC Genom Data. 2024 Jan 2;25(1):4. doi: 10.1186/s12863-023-01185-8. BMC Genom Data. 2024. PMID: 38166646 Free PMC article.
References
- Amorim A. Archaeogenetics. Journal of Iberian Archaeology. 1999;1:15–25.
- Richards M., Macaulay V., Bandelt H.-J. Analyzing genetic data in a model-based framework: Inferences about European prehistory. In: Bellwood P., Renfrew C., editors. Examining the language-farming dispersal hypothesis. McDonald Institute for Archaeological Research; Cambridge: 2002. pp. 459–466.
- Bandelt H.-J., Macaulay V., Richards M. What molecules can't tell us about the spread of languages and the Neolithic. In: Bellwood P., Renfrew C., editors. Examining the language-farming dispersal hypothesis. McDonald Institute for Archaeological Research; Cambridge: 2002. pp. 99–107.
- Macaulay V., Richards M. John Wiley & Sons, Ltd; Chichester: 2008. Mitochondrial genome sequences and their phylogeographic interpretation. Encyclopedia of Life Sciences (ELS)
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources