During muscle atrophy, thick, but not thin, filament components are degraded by MuRF1-dependent ubiquitylation - PubMed (original) (raw)
During muscle atrophy, thick, but not thin, filament components are degraded by MuRF1-dependent ubiquitylation
Shenhav Cohen et al. J Cell Biol. 2009.
Abstract
Loss of myofibrillar proteins is a hallmark of atrophying muscle. Expression of muscle RING-finger 1 (MuRF1), a ubiquitin ligase, is markedly induced during atrophy, and MuRF1 deletion attenuates muscle wasting. We generated mice expressing a Ring-deletion mutant MuRF1, which binds but cannot ubiquitylate substrates. Mass spectrometry of the bound proteins in denervated muscle identified many myofibrillar components. Upon denervation or fasting, atrophying muscles show a loss of myosin-binding protein C (MyBP-C) and myosin light chains 1 and 2 (MyLC1 and MyLC2) from the myofibril, before any measurable decrease in myosin heavy chain (MyHC). Their selective loss requires MuRF1. MyHC is protected from ubiquitylation in myofibrils by associated proteins, but eventually undergoes MuRF1-dependent degradation. In contrast, MuRF1 ubiquitylates MyBP-C, MyLC1, and MyLC2, even in myofibrils. Because these proteins stabilize the thick filament, their selective ubiquitylation may facilitate thick filament disassembly. However, the thin filament components decreased by a mechanism not requiring MuRF1.
Figures
Figure 1.
Targeted replacement of MuRF1 with Myc-MuRF1 or Myc-ΔR-MuRF1 and their expression in skeletal muscle after denervation. (A) Strategy for the generation of MuRF1_myc_/myc mice showing the Ring-finger domain (gray box) spanning the first two exons of MuRF1 (endogenous locus). The targeting cassette was generated by replacing the firsts two exons (E1 and E2) of MuRF1 with a Myc-encoding sequence fused in-frame with the region of MuRF1 encoding D2-S110, followed by a short intron sequence, and a selectable marker (NEO, neomycin resistance gene) (targeted locus). Upon homologous recombination, the floxed NEO cassette was removed with a Cre expression vector (recombined locus). (B) Immunoblot analysis of MuRF1 in gastrocnemius muscle from adult wild-type mice (WT) and Myc-MuRF1 from MuRF1_myc_/myc littermates after 10 d of denervation. The muscles on the right hindlimb were denervated by cutting the sciatic nerve, and the left hindlimb muscles of each animal served as the control. A MuRF1 antibody was used for Western blot. Asterisks indicate nonspecific bands. (C) Strategy for the generation of ΔR-MuRF1_myc_/myc mice showing the Ring-finger domain (gray box) spanning the first two exons of MuRF1 (endogenous locus). The targeting cassette was generated by replacing the first two exons (E1 and E2) of MuRF1 with a Myc-encoding sequence fused in-frame with the region of MuRF1 encoding D2-S110 without the sequence corresponding to the Ring-finger domain C23-C78, followed by a short intron sequence, and a selectable marker (NEO, neomycin resistance gene) (targeted locus). Upon homologous recombination, the floxed NEO gene was removed using a Cre expression vector (recombined locus). (D) Immunoblot analysis of MuRF1 in gastrocnemius muscle from WT mice and of ΔR-Myc-MuRF1 in muscles from ΔR-MuRF1_myc_/myc littermates 10 d after denervation. Antibodies used for Western blot are indicated. (E) Denervation atrophy of gastrocnemius in WT and MuRF1_myc_/myc or ΔR-MuRF1_myc_/myc mice. Weight of gastrocnemius in MuRF1myc/myc animals decreased 14 d after denervation to a similar extent as in WT littermates. By contrast, muscle weight from ΔR-MuRF1myc/myc animals decreased to a lesser extent than in WT littermates (*, P < 0.01). Weights of denervated muscles are expressed as percentage of the contralateral control. Error bars represent SEM (n = 7) versus innervated muscles from WT.
Figure 2.
MuRF1 is responsible for most of the increase in total ubiquitin conjugates during denervation atrophy. (A) The cytosolic fractions (20 µg) of 10-d denervated or control gastrocnemius muscles from MuRF1_myc/myc_ and ΔR-MuRF1_myc/myc_ mice were subjected to immunoblotting with an antibody against polyubiquitin conjugates. The membrane was re-probed with anti-actin to ensure equal protein loading. C, control; D, denervation for 10 d. (B) Densitometric analysis of Western blot presented in A, with mean given as arbitrary units. Error bars represent SEM, n = 4. #, P < 0.001 versus innervated muscles; *, P < 0.05 versus denervated muscles from ΔR-MuRF1_myc_/myc mice.
Figure 3.
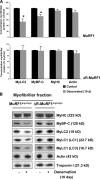
By 10 d denervation, MyLC1, MyLC2, and MyBP-C are selectively lost by a MuRF1-dependent mechanism. The myofibrillar fractions from 10-d denervated and control gastrocnemius muscles from both MuRF1_myc_/myc and ΔR-MuRF1_myc_/myc mice were analyzed by SDS-PAGE, Coomassie blue staining, and densitometric measurement of the specific bands. (A) The concentration of each protein per mg myofibrillar protein for denervated muscles is expressed as a percentage of the mean intensity in innervated controls. Error bars represent SEM, n = 3. *, P < 0.05 versus innervated muscles. (B) The myofibrillar fraction (0.5 µg) from denervated and innervated muscles from both types of mice was analyzed by SDS-PAGE and immunoblotted with specific antibodies as indicated.
Figure 4.
Pure MyLC1, MyLC2, MyBP-C, and MyHC are substrates of MuRF1, but in the actomyosin complex MyHC is not efficiently ubiquitylated. In vitro ubiquitylation of actomyosin complex or pure MyLC1 (A), MyLC2 (B), MyBP-C (C), and MyHC (D) by MuRF1 and three different E2s (UbcH13/Uev1). We assayed similar concentrations of pure MyHC and in association within actomyosin (as determined by Coomassie blue–stained gels). Asterisk indicates a nonspecific band. Dividing lines were used to separate top and bottom parts of the gel.
Figure 5.
MyBP-C and MyLCs are lost selectively during atrophy and are efficiently ubiquitylated by MuRF1 in isolated myofibrils. (A) Equal amounts of the myofibrillar fraction from innervated gastrocnemius muscles and ones denervated for 3 or 10 d from WT mice were analyzed by SDS-PAGE and Coomassie blue staining (left). The intensity of specific Coomassie blue–stained bands was measured, and the ratio to actin was plotted. Error bars represent SEM, n = 5. *, P < 0.05. Shown on the right is Western blot analysis of myofibrillar fraction (0.5 µg, to detect MyLC1) and cytosolic fractions (20 µg, to detect MuRF1) from innervated and denervated muscles from WT mice. (B) MyBP-C and MyLCs are ubiquitylated by purified MuRF1 in isolated myofibrils from control muscles and ones denervated for 3 or 10 d using UbcH1 and His-tagged ubiquitin. His-Ubiquitylated proteins were purified with a nickel column and detected by immunoblotting with specific antibodies.
Figure 6.
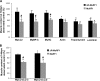
MyHC is lost from the myofibril by a MuRF1-dependent mechanism 14 d after denervation. The myofibrillar fraction from innervated muscles and ones denervated for 14 d from both MuRF1_myc/myc_ and ΔR-MuRF1_myc/myc_ mice were analyzed by SDS-PAGE, Coomassie blue staining (A), immunoblotting with specific antibodies as indicated (B), and densitometric measurement of the specific bands. The content of each myofibrillar protein in denervated muscle is presented as percentage of levels in control. Error bars represent SEM, n = 3. *, P < 0.05 denervation versus control. #, P < 0.05 denervation in MuRF1_myc_/myc versus ΔR-MuRF1_myc_/myc mice.
Similar articles
- Targeting MuRF1 to Combat Skeletal Muscle Wasting in Cardiac Cachexia: Mechanisms and Therapeutic Prospects.
Liu X, Wen Y, Lu Y. Liu X, et al. Med Sci Monit. 2024 Oct 22;30:e945211. doi: 10.12659/MSM.945211. Med Sci Monit. 2024. PMID: 39434377 Free PMC article. Review. - Muscle sparing in muscle RING finger 1 null mice: response to synthetic glucocorticoids.
Baehr LM, Furlow JD, Bodine SC. Baehr LM, et al. J Physiol. 2011 Oct 1;589(Pt 19):4759-76. doi: 10.1113/jphysiol.2011.212845. Epub 2011 Aug 1. J Physiol. 2011. PMID: 21807613 Free PMC article. - UBE2D2 is not involved in MuRF1-dependent muscle wasting during hindlimb suspension.
Polge C, Koulmann N, Claustre A, Jarzaguet M, Serrurier B, Combaret L, Béchet D, Bigard X, Attaix D, Taillandier D. Polge C, et al. Int J Biochem Cell Biol. 2016 Oct;79:488-493. doi: 10.1016/j.biocel.2016.06.019. Epub 2016 Jul 1. Int J Biochem Cell Biol. 2016. PMID: 27378730 - Targeting the ubiquitin E3 ligase MuRF1 to inhibit muscle atrophy.
Eddins MJ, Marblestone JG, Suresh Kumar KG, Leach CA, Sterner DE, Mattern MR, Nicholson B. Eddins MJ, et al. Cell Biochem Biophys. 2011 Jun;60(1-2):113-8. doi: 10.1007/s12013-011-9175-7. Cell Biochem Biophys. 2011. PMID: 21448668 - Signaling pathways perturbing muscle mass.
Glass DJ. Glass DJ. Curr Opin Clin Nutr Metab Care. 2010 May;13(3):225-9. doi: 10.1097/mco.0b013e32833862df. Curr Opin Clin Nutr Metab Care. 2010. PMID: 20397318 Review.
Cited by
- Targeting MuRF1 to Combat Skeletal Muscle Wasting in Cardiac Cachexia: Mechanisms and Therapeutic Prospects.
Liu X, Wen Y, Lu Y. Liu X, et al. Med Sci Monit. 2024 Oct 22;30:e945211. doi: 10.12659/MSM.945211. Med Sci Monit. 2024. PMID: 39434377 Free PMC article. Review. - Inhibition of nucleo-cytoplasmic proteasome translocation by the aromatic amino acids or silencing Sestrin3-their sensing mediator-is tumor suppressive.
Livneh I, Fabre B, Goldhirsh G, Lulu C, Zinger A, Shammai Vainer Y, Kaduri M, Dahan A, Ziv T, Schroeder A, Ben-Neriah Y, Zohar Y, Cohen-Kaplan V, Ciechanover A. Livneh I, et al. Cell Death Differ. 2024 Oct;31(10):1242-1254. doi: 10.1038/s41418-024-01370-x. Epub 2024 Sep 12. Cell Death Differ. 2024. PMID: 39266717 Free PMC article. - Proteasome gene expression is controlled by coordinated functions of multiple transcription factors.
Gilda JE, Nahar A, Kasiviswanathan D, Tropp N, Gilinski T, Lahav T, Alexandrovich D, Mandel-Gutfreund Y, Park S, Shemer S. Gilda JE, et al. J Cell Biol. 2024 Aug 5;223(8):e202402046. doi: 10.1083/jcb.202402046. Epub 2024 May 20. J Cell Biol. 2024. PMID: 38767572 - Murf1 alters myosin replacement rates in cultured myotubes in a myosin isoform-dependent manner.
Uenaka E, Ojima K, Suzuki T, Kobayashi K, Muroya S, Nishimura T. Uenaka E, et al. In Vitro Cell Dev Biol Anim. 2024 Aug;60(7):748-759. doi: 10.1007/s11626-024-00916-0. Epub 2024 May 17. In Vitro Cell Dev Biol Anim. 2024. PMID: 38758432 - Differential effects of cholecalciferol and calcitriol on muscle proteolysis and oxidative stress in angiotensin II-induced C2C12 myotube atrophy.
Hirunsai M, Srikuea R. Hirunsai M, et al. Physiol Rep. 2024 Apr;12(8):e16011. doi: 10.14814/phy2.16011. Physiol Rep. 2024. PMID: 38627219 Free PMC article.
References
- Bodine S.C., Latres E., Baumhueter S., Lai V.K., Nunez L., Clarke B.A., Poueymirou W.T., Panaro F.J., Na E., Dharmarajan K., et al. 2001. Identification of ubiquitin ligases required for skeletal muscle atrophy.Science. 294:1704–1708 - PubMed
- Centner T., Yano J., Kimura E., McElhinny A.S., Pelin K., Witt C.C., Bang M.L., Trombitas K., Granzier H., Gregorio C.C., et al. 2001. Identification of muscle specific ring finger proteins as potential regulators of the titin kinase domain.J. Mol. Biol. 306:717–726 - PubMed
- Clark K.A., McElhinny A.S., Beckerle M.C., Gregorio C.C. 2002. Striated muscle cytoarchitecture: an intricate web of form and function.Annu. Rev. Cell Dev. Biol. 18:637–706 - PubMed
- Clarke B.A., Drujan D., Willis M.S., Murphy L.O., Corpina R.A., Burova E., Rakhilin S.V., Stitt T.N., Patterson C., Latres E., Glass D.J. 2007. The E3 ligase MuRF1 degrades myosin heavy chain protein in dexamethasone-treated skeletal muscle.Cell Metab. 6:376–385 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases