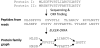An ORFome assembly approach to metagenomics sequences analysis - PubMed (original) (raw)
An ORFome assembly approach to metagenomics sequences analysis
Yuzhen Ye et al. J Bioinform Comput Biol. 2009 Jun.
Abstract
Metagenomics is an emerging methodology for the direct genomic analysis of a mixed community of uncultured microorganisms. The current analyses of metagenomics data largely rely on the computational tools originally designed for microbial genomics projects. The challenge of assembling metagenomic sequences arises mainly from the short reads and the high species complexity of the community. Alternatively, individual (short) reads will be searched directly against databases of known genes (or proteins) to identify homologous sequences. The latter approach may have low sensitivity and specificity in identifying homologous sequences, which may further bias the subsequent diversity analysis. In this paper, we present a novel approach to metagenomic data analysis, called Metagenomic ORFome Assembly (MetaORFA). The whole computational framework consists of three steps. Each read from a metagenomics project will first be annotated with putative open reading frames (ORFs) that likely encode proteins. Next, the predicted ORFs are assembled into a collection of peptides using an EULER assembly method. Finally, the assembled peptides (i.e. ORFome) are used for database searching of homologs and subsequent diversity analysis. We applied MetaORFA approach to several metagenomics datasets with low coverage short reads. The results show that MetaORFA can produce long peptides even when the sequence coverage of reads is extremely low. Hence, the ORFome assembly significantly increases the sensitivity of homology searching, and may potentially improve the diversity analysis of the metagenomic data. This improvement is especially useful for metagenomic projects when the genome assembly does not work because of the low sequence coverage.
Figures
Fig. 1
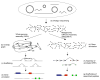
A schematic comparison of the ORFome assembly approach with the Whole Genome Assembly (WGA) pipeline for the metagenomic sequence analysis. Both approaches attempt to characterize the protein coding genes in the shotgun sequencing reads from the metagenomic analysis of an environmental sample containing a number of different microorganisms (the reads are shown as double-barreled, as currently several NGS techniques are capable of generating such data; however, some early metagnomics projects, including the datasets used in this paper, did not produce double-barreled sequencing reads, and thus the scaffolding step is not feasible) (a). The whole genome assembly (WGS) pipeline (b-d) first assembles the reads into contigs and scaffolds, and then annotates the genes in the assembled sequences. In comparison, ORFome assembly approach (e-g) first applies gene finding in the unassembled reads, and then assembles only those annotated (partial) ORFs into peptides. These peptides may be further connected to form scaffolds if there are mate-pairs available from double-barreled sequencing (g).
Fig. 2
A synthetic example for the ORFome assembly resulting into a protein family graph. Two homologous proteins are encoded in the metagenome. Due to the short read length, it is difficult to reconstruct the complete sequences of these two proteins. The EULER-ORFA approach assembles them into a protein family graph, in which the common and distinct parts between two proteins are represented by separate edges, and each protein corresponds to a path in the graph.
Fig. 3
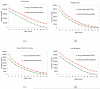
Comparison of the MetaORFA performance using different length cutoffs of input ORFs as shown in the total number of long assembled peptides (of at least 60aa)(a), and the length of the longest peptide (b).
Fig. 4
A long peptide with 155 aa (contig196081, highlighted in bold line) assembled from 18 putative ORFs (represented as thin lines below the contig) in the Gulf of Mexico dataset shows strong similarity with proteins in IMG database with known function (a). (b) shows the BLAST alignment between the peptide and the PhoH-like protein from Roseophage SIO1 in IMG database.
Fig. 5
Detailed comparison of the total number of read hits in IMG database using unassembled and the total number of read hits including those read hits belonging to the assembled peptides at different BLAST E-value cutoffs. The deviation between the two lines indicates the gain of read hits by using assembled peptides from the ORFome assembly.
Fig. 6
A peptide involving 11 synonymous polymorphic sites (starting from position 30, ending at position 60) assembled from the Sargasso Sea dataset. In the graph, the aligned protein sequences are shown on the top and the corresponding DNA sequences are shown on the bottom; the mutations are highlighted in bold and italic (if there are only two sequences covering the site, one arbitrary codon is highlighted).
Similar articles
- An ORFome assembly approach to metagenomics sequences analysis.
Ye Y, Tang H. Ye Y, et al. Comput Syst Bioinformatics Conf. 2008;7:3-13. Comput Syst Bioinformatics Conf. 2008. PMID: 19642264 - GRASPx: efficient homolog-search of short peptide metagenome database through simultaneous alignment and assembly.
Zhong C, Yang Y, Yooseph S. Zhong C, et al. BMC Bioinformatics. 2016 Aug 31;17 Suppl 8(Suppl 8):283. doi: 10.1186/s12859-016-1119-1. BMC Bioinformatics. 2016. PMID: 27585568 Free PMC article. - MetaDomain: a profile HMM-based protein domain classification tool for short sequences.
Zhang Y, Sun Y. Zhang Y, et al. Pac Symp Biocomput. 2012:271-82. Pac Symp Biocomput. 2012. PMID: 22174282 - Assessment of metagenomic assemblers based on hybrid reads of real and simulated metagenomic sequences.
Wang Z, Wang Y, Fuhrman JA, Sun F, Zhu S. Wang Z, et al. Brief Bioinform. 2020 May 21;21(3):777-790. doi: 10.1093/bib/bbz025. Brief Bioinform. 2020. PMID: 30860572 Free PMC article. Review. - A review of methods and databases for metagenomic classification and assembly.
Breitwieser FP, Lu J, Salzberg SL. Breitwieser FP, et al. Brief Bioinform. 2019 Jul 19;20(4):1125-1136. doi: 10.1093/bib/bbx120. Brief Bioinform. 2019. PMID: 29028872 Free PMC article. Review.
Cited by
- Computational meta'omics for microbial community studies.
Segata N, Boernigen D, Tickle TL, Morgan XC, Garrett WS, Huttenhower C. Segata N, et al. Mol Syst Biol. 2013 May 14;9:666. doi: 10.1038/msb.2013.22. Mol Syst Biol. 2013. PMID: 23670539 Free PMC article. Review. - Combining metagenomics, metatranscriptomics and viromics to explore novel microbial interactions: towards a systems-level understanding of human microbiome.
Bikel S, Valdez-Lara A, Cornejo-Granados F, Rico K, Canizales-Quinteros S, Soberón X, Del Pozo-Yauner L, Ochoa-Leyva A. Bikel S, et al. Comput Struct Biotechnol J. 2015 Jun 9;13:390-401. doi: 10.1016/j.csbj.2015.06.001. eCollection 2015. Comput Struct Biotechnol J. 2015. PMID: 26137199 Free PMC article. Review. - Novel sequence-based method for identifying transcription factor binding sites in prokaryotic genomes.
Sahota G, Stormo GD. Sahota G, et al. Bioinformatics. 2010 Nov 1;26(21):2672-7. doi: 10.1093/bioinformatics/btq501. Epub 2010 Aug 31. Bioinformatics. 2010. PMID: 20807838 Free PMC article. - Exploring the Human Microbiome: The Potential Future Role of Next-Generation Sequencing in Disease Diagnosis and Treatment.
Malla MA, Dubey A, Kumar A, Yadav S, Hashem A, Abd Allah EF. Malla MA, et al. Front Immunol. 2019 Jan 7;9:2868. doi: 10.3389/fimmu.2018.02868. eCollection 2018. Front Immunol. 2019. PMID: 30666248 Free PMC article. Review. - An Integrated Multi-Disciplinary Perspectivefor Addressing Challenges of the Human Gut Microbiome.
Shah RM, McKenzie EJ, Rosin MT, Jadhav SR, Gondalia SV, Rosendale D, Beale DJ. Shah RM, et al. Metabolites. 2020 Mar 6;10(3):94. doi: 10.3390/metabo10030094. Metabolites. 2020. PMID: 32155792 Free PMC article. Review.
References
- Galperin M. Metagenomics: from acid mine to shining sea. Environ Microbiol. 2004;6:543–545. - PubMed
- Eyers L, George I, Schuler L, et al. Environmental genomics: exploring the unmined richness of microbes to degrade xenobiotics. Appl Microbiol Biotechnol. 2004;66:123–130. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources