Cancer genome sequencing--an interim analysis - PubMed (original) (raw)
Cancer genome sequencing--an interim analysis
Edward J Fox et al. Cancer Res. 2009.
Abstract
With the publishing of the first complete whole genome of a human cancer and its paired normal, we have passed a key milestone in the cancer genome sequencing strategy. The generation of such data will, thanks to technical advances, soon become commonplace. As a significant number of proof-of-concept studies have been published, it is important to analyze now the likely implications of these data and how this information might frame cancer research in the near future. The diversity of genes mutated within individual tumor types, the most striking feature of all studies reported to date, challenges gene-centric models of tumorigenesis. Although cancer genome sequencing will revolutionize certain aspects of personalized care, the value of these studies in facilitating the development of new therapies, their primary goal, seems less promising. Most significantly, however, the cancer genome sequencing strategy, as currently applied, fails to characterize the most relevant genomic features of cancer-the mutational heterogeneity within individual tumors.
Figures
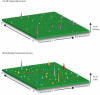
Figure 1. The Mutational Landscape of the Cancer Genome
(a) The cancer genome landscape of colorectal cancer as proposed by Wood et al. (9) demonstrates the mutational heterogeneity between individual tumors. The height of each violet peak represents the percentage of colorectal tumors found to carry a clonal mutation in a particular gene. The landscape comprises a small number of “mountains”, genes which are mutated in a large number of cancers [e.g. TP53 (23)], and a significantly larger number of “hills”–genes mutated in only a small number of individual tumors. While there may be 50 or more genes clonally mutated in a given tumor, any given gene is rarely mutated in more than a few tumors. (b) The mutational heterogeneity within individual tumors not detected by the cancer genome sequencing studies. The mutational landscape of three tumors are shown; each tumor is represented by a set of differently colored peaks (blue, red and yellow). The height of each peak reflects the number of cells within a single tumor that have a particular mutated gene. Split colored peaks represent the small number of mutated genes shared in common between more than one tumor. Within an individual neoplasm, a few mutations are present throughout the population (tall peaks), a greater number are present in minority subclones (short peaks) and the majority are found in only one or a few cells (invisibly distributed throughout the green background.)
Similar articles
- [Cancer genome analysis through next-generation sequencing].
Aburatani H. Aburatani H. Gan To Kagaku Ryoho. 2011 Jan;38(1):1-6. Gan To Kagaku Ryoho. 2011. PMID: 21368453 Review. Japanese. - A 2015 update on predictive molecular pathology and its role in targeted cancer therapy: a review focussing on clinical relevance.
Dietel M, Jöhrens K, Laffert MV, Hummel M, Bläker H, Pfitzner BM, Lehmann A, Denkert C, Darb-Esfahani S, Lenze D, Heppner FL, Koch A, Sers C, Klauschen F, Anagnostopoulos I. Dietel M, et al. Cancer Gene Ther. 2015 Sep;22(9):417-30. doi: 10.1038/cgt.2015.39. Epub 2015 Sep 11. Cancer Gene Ther. 2015. PMID: 26358176 Review. - Novel clinico-genome network modeling for revolutionizing genotype-phenotype-based personalized cancer care.
Roukos DH. Roukos DH. Expert Rev Mol Diagn. 2010 Jan;10(1):33-48. doi: 10.1586/erm.09.69. Expert Rev Mol Diagn. 2010. PMID: 20014921 Review. - Comprehensive next-generation cancer genome sequencing in the era of targeted therapy and personalized oncology.
Cronin M, Ross JS. Cronin M, et al. Biomark Med. 2011 Jun;5(3):293-305. doi: 10.2217/bmm.11.37. Biomark Med. 2011. PMID: 21657839 Review.
Cited by
- Clonal expansions in ulcerative colitis identify patients with neoplasia.
Salk JJ, Salipante SJ, Risques RA, Crispin DA, Li L, Bronner MP, Brentnall TA, Rabinovitch PS, Horwitz MS, Loeb LA. Salk JJ, et al. Proc Natl Acad Sci U S A. 2009 Dec 8;106(49):20871-6. doi: 10.1073/pnas.0909428106. Epub 2009 Nov 19. Proc Natl Acad Sci U S A. 2009. PMID: 19926851 Free PMC article. - Can we deconstruct cancer, one patient at a time?
Blau CA, Liakopoulou E. Blau CA, et al. Trends Genet. 2013 Jan;29(1):6-10. doi: 10.1016/j.tig.2012.09.004. Epub 2012 Oct 23. Trends Genet. 2013. PMID: 23102584 Free PMC article. - Cancer systems biology: a network modeling perspective.
Kreeger PK, Lauffenburger DA. Kreeger PK, et al. Carcinogenesis. 2010 Jan;31(1):2-8. doi: 10.1093/carcin/bgp261. Epub 2009 Oct 27. Carcinogenesis. 2010. PMID: 19861649 Free PMC article. Review. - Human cancers express mutator phenotypes: origin, consequences and targeting.
Loeb LA. Loeb LA. Nat Rev Cancer. 2011 Jun;11(6):450-7. doi: 10.1038/nrc3063. Epub 2011 May 19. Nat Rev Cancer. 2011. PMID: 21593786 Free PMC article. Review. - Active site mutations in mammalian DNA polymerase delta alter accuracy and replication fork progression.
Schmitt MW, Venkatesan RN, Pillaire MJ, Hoffmann JS, Sidorova JM, Loeb LA. Schmitt MW, et al. J Biol Chem. 2010 Oct 15;285(42):32264-72. doi: 10.1074/jbc.M110.147017. Epub 2010 Jul 13. J Biol Chem. 2010. PMID: 20628184 Free PMC article.
References
- Weinberg RA. The Biology of Cancer. Garland Science. 2006
- Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61:759–67. - PubMed
- Grady WM, Markowitz SD. Genetic and epigenetic alterations in colon cancer. Annu Rev Genomics Hum Genet. 2002;3:101–28. - PubMed
- Pollack A. New York Times. 2005 March 28; Sect. A1.
Publication types
MeSH terms
Grants and funding
- AG033485/AG/NIA NIH HHS/United States
- F30 AG033485-01/AG/NIA NIH HHS/United States
- R01 CA102029-07/CA/NCI NIH HHS/United States
- R01 CA115802/CA/NCI NIH HHS/United States
- R01 CA078885/CA/NCI NIH HHS/United States
- CA78885/CA/NCI NIH HHS/United States
- R01 CA102029/CA/NCI NIH HHS/United States
- R01 CA115802-02/CA/NCI NIH HHS/United States
- CA102029/CA/NCI NIH HHS/United States
- F30 AG033485/AG/NIA NIH HHS/United States
LinkOut - more resources
Full Text Sources
Research Materials