Extensive demethylation of repetitive elements during seed development underlies gene imprinting - PubMed (original) (raw)
Extensive demethylation of repetitive elements during seed development underlies gene imprinting
Mary Gehring et al. Science. 2009.
Abstract
DNA methylation is an epigenetic mark associated with transposable element silencing and gene imprinting in flowering plants and mammals. In plants, imprinting occurs in the endosperm, which nourishes the embryo during seed development. We have profiled Arabidopsis DNA methylation genome-wide in the embryo and endosperm and found that large-scale methylation changes accompany endosperm development and endosperm-specific gene expression. Transposable element fragments are extensively demethylated in the endosperm. We discovered new imprinted genes by the identification of candidates associated with regions of reduced endosperm methylation and preferential expression in endosperm relative to other parts of the plant. These data suggest that imprinting in plants evolved from targeted methylation of transposable element insertions near genic regulatory elements followed by positive selection when the resulting expression change was advantageous.
Figures
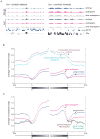
Figure 1. Endosperm is less methylated than embryo
A) Typical methylation profiles from a gene-rich and gene-poor region of the genome showing highly similar methylation patterns in embryo, endosperm, and dme endosperm from the L_er_ and Col-gl accessions. B) Average methylation in 100-bp windows of protein-coding genes and transposable element genes aligned at either their 5′ or 3′ ends in Col-gl embryo and endosperm. C) Methylation profiles of protein-coding genes when a set of 31,076 transposable element fragments (11) are and are not excluded from the averaging.
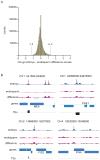
Figure 2. Known imprinted genes are associated with top DMRs
A) Histogram of Col-gl embryo - endosperm difference scores for 1.2 million overlapping 300-bp segments. Dashed lines represent the cutoff for the top 0.5% of methylation differences (~6000 300-bp segments). B) Embryo and endosperm methylation profiles of known imprinted genes. Red arrows, regions within the top 0.5% of methylation differences; gray arrow, region below the cutoff.
Figure 3. Methylation is lost 5′ and 3′ of genes in the endosperm
Average methylation profiles in embryo, endosperm, and dme endosperm of the A) 1276 Col-gl and B) 1163 Ler genes associated with more methylation in embryo than endosperm. Genes were aligned at their 5′ or 3′ end and the average methylation determined every 100 bp.
Figure 4. Expression and methylation analysis of new imprinted genes
A) RT-PCR allele-specific expression analysis from endosperm RNA of Col-gl females X L_er_ males and L_er_ females X Col-gl males. FWA is a control imprinted gene; AT3G25260 is biallelic. B) RT-PCR sequencing chromatograms for the paternally expressed genes AT2G32370 and AT5G62110. The biallelically-expressed gene αVPE is shown as a control. C) and D) Methylation profiling and allele-specific bisulfite sequencing analysis of new imprinted genes. Dashed lines represent the cutoff for the top 0.5% of differences. Each line of circles represent a bisulfite sequencing clone from embryo or endosperm. Filled circle, methylated cytosine. Red, CG; blue, CHG; gray, CHH.
Similar articles
- Genome-wide demethylation of Arabidopsis endosperm.
Hsieh TF, Ibarra CA, Silva P, Zemach A, Eshed-Williams L, Fischer RL, Zilberman D. Hsieh TF, et al. Science. 2009 Jun 12;324(5933):1451-4. doi: 10.1126/science.1172417. Science. 2009. PMID: 19520962 Free PMC article. - Genomic imprinting: insights from plants.
Gehring M. Gehring M. Annu Rev Genet. 2013;47:187-208. doi: 10.1146/annurev-genet-110711-155527. Epub 2013 Aug 30. Annu Rev Genet. 2013. PMID: 24016190 Review. - Genomic analysis of parent-of-origin allelic expression in Arabidopsis thaliana seeds.
Gehring M, Missirian V, Henikoff S. Gehring M, et al. PLoS One. 2011;6(8):e23687. doi: 10.1371/journal.pone.0023687. Epub 2011 Aug 17. PLoS One. 2011. PMID: 21858209 Free PMC article. - Paternally Acting Canonical RNA-Directed DNA Methylation Pathway Genes Sensitize Arabidopsis Endosperm to Paternal Genome Dosage.
Satyaki PRV, Gehring M. Satyaki PRV, et al. Plant Cell. 2019 Jul;31(7):1563-1578. doi: 10.1105/tpc.19.00047. Epub 2019 May 7. Plant Cell. 2019. PMID: 31064867 Free PMC article. - [Genomic imprinting and seed development].
Zhang WW, Cao SX, Jiang L, Zhu SS, Wan JM. Zhang WW, et al. Yi Chuan. 2005 Jul;27(4):665-70. Yi Chuan. 2005. PMID: 16120596 Review. Chinese.
Cited by
- Involvement of 5mC DNA demethylation via 5-aza-2'-deoxycytidine in regulating gene expression during early somatic embryo development in white spruce (Picea glauca).
Gao Y, Chen X, Liu C, Zhao H, Dai F, Zhao J, Zhang J, Kong L. Gao Y, et al. For Res (Fayettev). 2023 Dec 26;3:30. doi: 10.48130/fr-0023-0030. eCollection 2023. For Res (Fayettev). 2023. PMID: 39526256 Free PMC article. - Unveiling the imprinted dance: how parental genomes orchestrate seed development and hybrid success.
Muthusamy M, Pandian S, Shin EK, An HK, Sohn SI. Muthusamy M, et al. Front Plant Sci. 2024 Sep 27;15:1455685. doi: 10.3389/fpls.2024.1455685. eCollection 2024. Front Plant Sci. 2024. PMID: 39399543 Free PMC article. Review. - Epigenetic and transcriptional consequences in the endosperm of chemically induced transposon mobilization in Arabidopsis.
Del Toro-De León G, van Boven J, Santos-González J, Jiao WB, Peng H, Schneeberger K, Köhler C. Del Toro-De León G, et al. Nucleic Acids Res. 2024 Aug 27;52(15):8833-8848. doi: 10.1093/nar/gkae572. Nucleic Acids Res. 2024. PMID: 38967011 Free PMC article. - A DNA demethylase reduces seed size by decreasing the DNA methylation of AT-rich transposable elements in soybean.
Wang W, Zhang T, Liu C, Liu C, Jiang Z, Zhang Z, Ali S, Li Z, Wang J, Sun S, Chen Q, Zhang Q, Xie L. Wang W, et al. Commun Biol. 2024 May 21;7(1):613. doi: 10.1038/s42003-024-06306-2. Commun Biol. 2024. PMID: 38773248 Free PMC article. - United by conflict: Convergent signatures of parental conflict in angiosperms and placental mammals.
Soliman HK, Coughlan JM. Soliman HK, et al. J Hered. 2024 Oct 23;115(6):625-642. doi: 10.1093/jhered/esae009. J Hered. 2024. PMID: 38366852 Review.
References
- Huh JH, Bauer MJ, Hsieh TF, Fischer RL. Cell. 2008;132:735. - PubMed
- Gutierrez-Marcos JF, et al. Nat Genet. 2006;38:876. - PubMed
- Zilberman D, Gehring M, Tran RK, Ballinger T, Henikoff S. Nat Genet. 2007;39:61. - PubMed
- Zhang X, et al. Cell. 2006;126:1189. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources