Genome-wide demethylation of Arabidopsis endosperm - PubMed (original) (raw)
Genome-wide demethylation of Arabidopsis endosperm
Tzung-Fu Hsieh et al. Science. 2009.
Abstract
Parent-of-origin-specific (imprinted) gene expression is regulated in Arabidopsis thaliana endosperm by cytosine demethylation of the maternal genome mediated by the DNA glycosylase DEMETER, but the extent of the methylation changes is not known. Here, we show that virtually the entire endosperm genome is demethylated, coupled with extensive local non-CG hypermethylation of small interfering RNA-targeted sequences. Mutation of DEMETER partially restores endosperm CG methylation to levels found in other tissues, indicating that CG demethylation is specific to maternal sequences. Endosperm demethylation is accompanied by CHH hypermethylation of embryo transposable elements. Our findings demonstrate extensive reconfiguration of the endosperm methylation landscape that likely reinforces transposon silencing in the embryo.
Figures
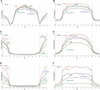
Fig. 1
Profiles of DNA methylation in embryo, wild-type endosperm, and dme endosperm. (A to F) TAIR8-annotated genes [(A), (C), and (E)] or transposons [(B), (D), and (F)] were aligned at the 5′ end (left panel) or the 3′ end (right panel), and average methylation levels for each 100-bp interval are plotted from 2 kb away from the gene (negative numbers) to 4 kb into the gene (positive numbers). Embryo methylation is represented by the red trace, wild-type (WT) endosperm by the blue trace, dme endosperm by the green trace, and aerial tissues by the black trace. The dashed line at zero represents the point of alignment. CG methylation is shown in (A) and (B), CHG in (C) and (D), CHH in (E) and (F).
Fig. 2
Associations between endosperm methylation, siRNAs, and expression. (A) Box plots showing siRNA abundance within 50-bp windows in the entire Arabidopsis genome (All) and in sequences hypermethylated in WT endosperm compared with the embryo in the CHG and CHH contexts. (B) Box plots showing differences in gene expression between embryo and endosperm for all genes (n = 21,021), genes with 5′ hypomethylation in endosperm (n = 1097), and genes with 3′ hypomethylation in endosperm (n = 505). Each box encloses the middle 50% of the distribution, with the horizontal line marking the median and the dot marking the mean. The lines extending from each box mark the minimum and maximum values that fall within 1.5 times the height of the box.
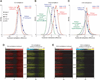
Fig. 3
Genome-wide demethylation of endosperm. (A to C) Kernel density plots of the differences between embryo and WT endosperm methylation (blue trace) and the differences between embryo and dme endosperm methylation (red trace). The green trace in (B) and (C) represents methylation differences between embryo and dme endosperm for windows with absolute fractional methylation increase in WT endosperm compared with embryo of at least 0.4 in the CHG context (B) (n = 135) or at least 0.2 in the CHH context (C) (n = 6168). Methylation differences for the 3′ MEA repeats, FWA, FIS2, PHE1, and MPC are indicated; specifics are listed in table S2. (D and E) All TAIR8-annotated genes (28,244) were aligned at the 5′ end and stacked from the top of chromosome 1 to the bottom of chromosome 5. Embryo methylation is displayed as a heat map in the left panel, differences between embryo and WT endosperm in the right panel. CG methylation is shown in (D), CHG in (E).
Similar articles
- Extensive demethylation of repetitive elements during seed development underlies gene imprinting.
Gehring M, Bubb KL, Henikoff S. Gehring M, et al. Science. 2009 Jun 12;324(5933):1447-51. doi: 10.1126/science.1171609. Science. 2009. PMID: 19520961 Free PMC article. - Active DNA demethylation in plant companion cells reinforces transposon methylation in gametes.
Ibarra CA, Feng X, Schoft VK, Hsieh TF, Uzawa R, Rodrigues JA, Zemach A, Chumak N, Machlicova A, Nishimura T, Rojas D, Fischer RL, Tamaru H, Zilberman D. Ibarra CA, et al. Science. 2012 Sep 14;337(6100):1360-1364. doi: 10.1126/science.1224839. Science. 2012. PMID: 22984074 Free PMC article. - One-way control of FWA imprinting in Arabidopsis endosperm by DNA methylation.
Kinoshita T, Miura A, Choi Y, Kinoshita Y, Cao X, Jacobsen SE, Fischer RL, Kakutani T. Kinoshita T, et al. Science. 2004 Jan 23;303(5657):521-3. doi: 10.1126/science.1089835. Epub 2003 Nov 20. Science. 2004. PMID: 14631047 - Genome demethylation and imprinting in the endosperm.
Bauer MJ, Fischer RL. Bauer MJ, et al. Curr Opin Plant Biol. 2011 Apr;14(2):162-7. doi: 10.1016/j.pbi.2011.02.006. Epub 2011 Mar 23. Curr Opin Plant Biol. 2011. PMID: 21435940 Free PMC article. Review. - Imprinting in plants and its underlying mechanisms.
Zhang H, Chaudhury A, Wu X. Zhang H, et al. J Genet Genomics. 2013 May 20;40(5):239-47. doi: 10.1016/j.jgg.2013.04.003. Epub 2013 Apr 20. J Genet Genomics. 2013. PMID: 23706299 Review.
Cited by
- Utilizing Short Interspersed Nuclear Element as a Genetic Marker for Pre-Harvest Sprouting in Wheat.
Kandpal P, Kaur K, Dhariwal R, Kaur S, Brar GK, Randhawa H, Singh J. Kandpal P, et al. Plants (Basel). 2024 Oct 25;13(21):2981. doi: 10.3390/plants13212981. Plants (Basel). 2024. PMID: 39519902 Free PMC article. - Upstream regulator of genomic imprinting in rice endosperm is a small RNA-associated chromatin remodeler.
Pal AK, Gandhivel VH, Nambiar AB, Shivaprasad PV. Pal AK, et al. Nat Commun. 2024 Sep 6;15(1):7807. doi: 10.1038/s41467-024-52239-z. Nat Commun. 2024. PMID: 39242590 Free PMC article. - United by conflict: Convergent signatures of parental conflict in angiosperms and placental mammals.
Soliman HK, Coughlan JM. Soliman HK, et al. J Hered. 2024 Oct 23;115(6):625-642. doi: 10.1093/jhered/esae009. J Hered. 2024. PMID: 38366852 Review. - Molecular basis and evolutionary drivers of endosperm-based hybridization barriers.
Bente H, Köhler C. Bente H, et al. Plant Physiol. 2024 Apr 30;195(1):155-169. doi: 10.1093/plphys/kiae050. Plant Physiol. 2024. PMID: 38298124 Free PMC article. Review. - H2A.X promotes endosperm-specific DNA methylation in Arabidopsis thaliana.
Frost JM, Lee J, Hsieh PH, Lin SJH, Min Y, Bauer M, Runkel AM, Cho HT, Hsieh TF, Fischer RL, Choi Y. Frost JM, et al. BMC Plant Biol. 2023 Nov 22;23(1):585. doi: 10.1186/s12870-023-04596-y. BMC Plant Biol. 2023. PMID: 37993808 Free PMC article.
References
- Feil R, Berger F. Trends Genet. 2007;23:192. - PubMed
- Choi Y, et al. Cell. 2002;110:33. - PubMed
- Huh JH, Bauer MJ, Hsieh T-F, Fischer RL. Cell. 2008;132:735. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases