Complex adaptations can drive the evolution of the capacitor [PSI], even with realistic rates of yeast sex - PubMed (original) (raw)
Complex adaptations can drive the evolution of the capacitor [PSI], even with realistic rates of yeast sex
Cortland K Griswold et al. PLoS Genet. 2009 Jun.
Abstract
The [PSI(+)] prion may enhance evolvability by revealing previously cryptic genetic variation, but it is unclear whether such evolvability properties could be favored by natural selection. Sex inhibits the evolution of other putative evolvability mechanisms, such as mutator alleles. This paper explores whether sex also prevents natural selection from favoring modifier alleles that facilitate [PSI(+)] formation. Sex may permit the spread of "cheater" alleles that acquire the benefits of [PSI(+)] through mating without incurring the cost of producing [PSI(+)] at times when it is not adaptive. Using recent quantitative estimates of the frequency of sex in Saccharomyces paradoxus, we calculate that natural selection for evolvability can drive the evolution of the [PSI(+)] system, so long as yeast populations occasionally require complex adaptations involving synergistic epistasis between two loci. If adaptations are always simple and require substitution at only a single locus, then the [PSI(+)] system is not favored by natural selection. Obligate sex might inhibit the evolution of [PSI(+)]-like systems in other species.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
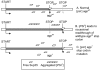
Figure 1. Different mechanisms of readthrough translation.
Either the presence of [_PSI+_] (B) or an agp + point mutation (C) can lead to readthrough of the wild-type stop codon (A).
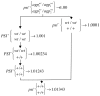
Figure 2. Alternative pathways, with and without [_PSI+_], leading to the same readthrough adaptation.
Adaptation in environment 2 can proceed either via: (1) [_PSI+_] appearance followed by point mutation at agp loci (genetic assimilation) and finally reversion to [psi −] (left) and (2) direct adaptation at agp loci without involvement of [_PSI+_] (right). The fitness of an individual is given to the right of its genotype, calculated using s 2 = 0.001. Only homozygous states are shown because inbreeding quickly leads to homozygosity. The genetic assimilation pathway typically occurs more often because [_PSI+_] individuals appear far more often than [_psi−_] individuals who carry the “+” allele at both agp loci.
Figure 3. An example of two-locus adaptation mediated by [_PSI+_].
ε = 0.01, Ω12 = Ω21 = 10−5, s 2 = 0.001, h = 1.
Figure 4. prf+ is maintained in the two-locus but not the one-locus model.
The _y_-axis gives the probability that the frequency of prf+ after 5×105 generations is greater than its starting frequency of 0.5. The strength of selection s2 for adaptation in environment 2 affects the cutoff frequency of sex. ε = 0.01, Ω12 = Ω21 = 10−5. prf+ is maintained in the two-locus model unless sex is very frequent.
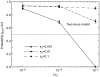
Figure 5. If environment 2 is too short-lived (high Ω21), then prf+ is not maintained.
This is because there is insufficient time for the selective sweeps shown in Figure 3 to be completed. ε = 0.01, Ω12 = 10−5, h = 1.
Figure 6. Very rare opportunities for adaptation (low Ω12) cause prf+ to be lost.
ε = 0.01, Ω21 = 10−5.
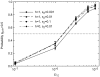
Figure 7. Fixation probabilities starting from a single copy.
prf+ fixes more often than the neutral expectation (10−7), except when selection is weak (s 2≤0.001) and sex is common _psex_≥0.1. All parameters are equal to values in Figure 3, with the additional parameter μprf+ equal to 10−9. For psex = 0.001 and psex = 0.01, results are based on 106 replicates. For psex = 0.1 and s2 = 0.001, the number of replicates is 3.5×107. Otherwise the number of replicates is 6×106.
Figure 8. Fixation probabilities starting from a single copy.
prf+ allele fixes above neutral expectations (10−7) when the transition probability to environment 2 is greater than 10−8. All parameters are equal to values in Figure 6, with the additional parameter μprf+ = 10−9. Results are based on 106 replicates (Ω12 = 10−5), 5×106 replicates (Ω12 = 10−6), 2×107 replicates (Ω12 = 10−7) and 2×107 replicates (Ω12 = 10−8).
Similar articles
- The strength of selection against the yeast prion [PSI+].
Masel J, Griswold CK. Masel J, et al. Genetics. 2009 Mar;181(3):1057-63. doi: 10.1534/genetics.108.100297. Epub 2009 Jan 19. Genetics. 2009. PMID: 19153253 Free PMC article. - The evolution of the evolvability properties of the yeast prion [PSI+].
Masel J, Bergman A. Masel J, et al. Evolution. 2003 Jul;57(7):1498-512. doi: 10.1111/j.0014-3820.2003.tb00358.x. Evolution. 2003. PMID: 12940355 - Prion switching in response to environmental stress.
Tyedmers J, Madariaga ML, Lindquist S. Tyedmers J, et al. PLoS Biol. 2008 Nov 25;6(11):e294. doi: 10.1371/journal.pbio.0060294. PLoS Biol. 2008. PMID: 19067491 Free PMC article. - The three faces of Sup35.
Lyke DR, Dorweiler JE, Manogaran AL. Lyke DR, et al. Yeast. 2019 Aug;36(8):465-472. doi: 10.1002/yea.3392. Epub 2019 Jul 1. Yeast. 2019. PMID: 30963611 Free PMC article. Review. - [New aspects of research upon the yeast Saccharomyces cerevisiae [PSI+] prion].
Ishikawa T. Ishikawa T. Postepy Biochem. 2007;53(2):182-7. Postepy Biochem. 2007. PMID: 17969880 Review. Polish.
Cited by
- Mistranslation can enhance fitness through purging of deleterious mutations.
Bratulic S, Toll-Riera M, Wagner A. Bratulic S, et al. Nat Commun. 2017 May 19;8:15410. doi: 10.1038/ncomms15410. Nat Commun. 2017. PMID: 28524864 Free PMC article. - Prions are a common mechanism for phenotypic inheritance in wild yeasts.
Halfmann R, Jarosz DF, Jones SK, Chang A, Lancaster AK, Lindquist S. Halfmann R, et al. Nature. 2012 Feb 15;482(7385):363-8. doi: 10.1038/nature10875. Nature. 2012. PMID: 22337056 Free PMC article. - Protein self-assembly: A new frontier in cell signaling.
Saad S, Jarosz DF. Saad S, et al. Curr Opin Cell Biol. 2021 Apr;69:62-69. doi: 10.1016/j.ceb.2020.12.013. Epub 2021 Jan 23. Curr Opin Cell Biol. 2021. PMID: 33493989 Free PMC article. Review. - On the Nature and Evolutionary Impact of Phenotypic Robustness Mechanisms.
Siegal ML, Leu JY. Siegal ML, et al. Annu Rev Ecol Evol Syst. 2014 Nov 1;45:496-517. doi: 10.1146/annurev-ecolsys-120213-091705. Annu Rev Ecol Evol Syst. 2014. PMID: 26034410 Free PMC article. - Assessment of inactivating stop codon mutations in forty Saccharomyces cerevisiae strains: implications for [PSI] prion- mediated phenotypes.
Fitzpatrick DA, O'Brien J, Moran C, Hasin N, Kenny E, Cormican P, Gates A, Morris DW, Jones GW. Fitzpatrick DA, et al. PLoS One. 2011;6(12):e28684. doi: 10.1371/journal.pone.0028684. Epub 2011 Dec 15. PLoS One. 2011. PMID: 22194885 Free PMC article.
References
- Wickner RB, Masison DC, Edskes HK. [PSI] and [URE3] as yeast prions. Yeast. 1995;11:1671–1685. - PubMed
- Lund PM, Cox BS. Reversion analysis of [psi −] mutations in Saccharomyces cerevisiae. Genet Res. 1981;37:173–182. - PubMed
- Serio TR, Lindquist SL. [PSI+]: An epigenetic modulator of translation termination efficiency. Annu Rev Cell Dev Biol. 1999;15:661–703. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases