H3 k36 methylation helps determine the timing of cdc45 association with replication origins - PubMed (original) (raw)
H3 k36 methylation helps determine the timing of cdc45 association with replication origins
Fiona Pryde et al. PLoS One. 2009.
Abstract
Background: Replication origins fire at different times during S-phase. Such timing is determined by the chromosomal context, which includes the activity of nearby genes, telomeric position effects and chromatin structure, such as the acetylation state of the surrounding chromatin. Activation of replication origins involves the conversion of a pre-replicative complex to a replicative complex. A pivotal step during this conversion is the binding of the replication factor Cdc45, which associates with replication origins at approximately their time of activation in a manner partially controlled by histone acetylation.
Methodology/principal findings: Here we identify histone H3 K36 methylation (H3 K36me) by Set2 as a novel regulator of the time of Cdc45 association with replication origins. Deletion of SET2 abolishes all forms of H3 K36 methylation. This causes a delay in Cdc45 binding to origins and renders the dynamics of this interaction insensitive to the state of histone acetylation of the surrounding chromosomal region. Furthermore, a decrease in H3 K36me3 and a concomitant increase in H3 K36me1 around the time of Cdc45 binding to replication origins suggests opposing functions for these two methylation states. Indeed, we find K36me3 depleted from early firing origins when compared to late origins genomewide, supporting a delaying effect of this histone modification for the association of replication factors with origins.
Conclusions/significance: We propose a model in which K36me1 together with histone acetylation advance, while K36me3 and histone deacetylation delay, the time of Cdc45 association with replication origins. The involvement of the transcriptionally induced H3 K36 methylation mark in regulating the timing of Cdc45 binding to replication origins provides a novel means of how gene expression may affect origin dynamics during S-phase.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
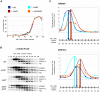
Figure 1. H3 K36me by Set2 is necessary for accelerated S-phase progression in Δrpd3 cells.
(A) Exponentially growing cells of strains MVY17 (WT), MVY31 (Δ_rpd3_), MVY37 (K36/37R) and MVY34 (Δ_rpd3_K36/37R) were arrested in G1 with α-factor and released into S-phase at 30°C. Samples were taken at indicated times and processed for FACS analysis. G1 and G2 DNA content are indicated at the bottom of the figure. Grey bars indicate the estimated length of S-phase. (B) Same as (A) but with strains MMY001 (WT), MMY002 (Δ_rpd3_), MVY42 (Δ_set2_) and MVY43 (Δ_rpd3_Δ_set2_).
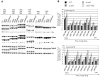
Figure 2. Set2p is necessary for advanced association of Cdc45 with origins in WT and Δrpd3 cells.
Strains MMY033 (WT), MVY51 (Δ_rpd3_), MVY57 (Δ_set2_) and MVY58 (Δ_rpd3_Δ_set2_) were arrested in G1 with α-factor, released at 24°C into S-phase and samples were taken at indicated times. (A) Cell budding was assessed by microscopy. (B) ChIP of Cdc45-3FLAG was performed with an α-FLAG antibody and analysed by semiquantitative PCR using primers specific for ARS607, ARS1412 and a telomeric loading control (TEL). (C) Graphical representation of Cdc45-3FLAG ChIP showing the relative intensity of ARS-specific fragments after normalization to the telomeric loading control fragment and the input DNA. The time of maximal intensity is indicated for each strain.
Figure 3. Δset2 does not decrease histone acetylation at origins.
ChIP of MMY033 (WT), MVY51 (Δ_rpd3_), MVY57 (Δ_set2_) and MVY58 (Δ_rpd3_Δ_set2_) with antibodies specific for pan-acetylated histone H3 and H4 was performed. (A) Representative autoradiographs of PCR products using primer pairs specific for the indicated ARS elements and a telomeric loading control (TEL). (B) Graphic representation: Analysis was by semiquantitative PCR using primer pairs specific for indicated ARS elements and a telomeric loading control. The relative intensity of ARS specific fragments after normalization to the loading control and the input is presented. Errorbars refer to the standard deviation of the results of three independent experiments.
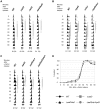
Figure 4. Δrpd3 increases K36me at some but not all origins.
ChIP of MMY033 (WT) and MVY51 (Δrpd3) with antibodies specific for K36me1 (A) and −me3 (B). ChIP of strain MVY57 (Δ_set2_) is included as a negative control in (A). Representative autoradiographs of PCR products using primers specific for the indicated ARS elements and a telomeric loading control (TEL). The graph represents the average ratio (Δ_rpd3_/WT) of three independent experiments after normalization to input DNA and loading control. Error-bars refer to the standard deviations thereof.
Figure 5. K36me1 and K36me3 show opposing behaviour at DNA replication origins.
(A) The ratio of K36me3/K36me1 for indicated ARSs is plotted against their time of activation and a linear trend-line was drawn. (B) ARSs are grouped for their time of firing in three minutes time intervals relative to the firing of the earliest ARS . ChIP on CHIP (Nimblegen) was used to determine genome-wide levels of H3 K36me3 and their distribution was determined for each group of ARSs and represented as box-plot. W- and P-values described in the text were calculated using the Wilcoxon rank sum test with continuity correction. (C) Same as in (B), but excluding all ARSs within 30 kb from chromosomal ends. (D) ChIP of MMY033 (WT) and MVY51 (Δ_rpd3_) with antibodies specific for K36me1 and −me3. Representative autoradiographs of PCR products using primers specific for the indicated ARS elements and a telomeric loading control (TEL). (E) Graphical representation of K36me1 or K36me3 ChIP showing the relative intensity of ARS-specific fragments after normalization to the loading control and the input. Complete lines indicate WT and broken lines indicate Δ_rpd3_.
Figure 6. EAF3 and NTO1 act together to delay S-phase progression.
(A) Strains MMY033 (WT), MVY51 (Δ_rpd3_), MVY54 (Δ_eaf3_) and MVY55 (Δ_eaf3_Δ_rpd3_) were arrested in G1 with α-factor and released into S-phase at 30°C. Samples were taken at indicated times and processed for FACS analysis. (B) as (A) with strains MMY001 (WT), MMY002 (Δ_rpd3_), MMY118 (Δ_nto1_) and MVY119 (Δ_rpd3_Δ_nto1_). (C) as (A) with strains MMY001 (WT), MMY002 (Δrpd3), MVY137 (Δ_eaf3_Δ_nto1_) and MVY138 (Δ_rpd3_Δ_eaf3_Δ_nto1_). Grey bars indicate the estimated length of S-phase. (D) Cell budding was assessed by microscopy. Complete lines indicate WT (black) and Δ_rpd3_ (grey); broken lines indicate Δ_eaf3_Δ_nto1_ (black) and Δ_rpd3_Δ_eaf3_Δ_nto1_ (grey).
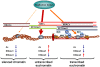
Figure 7. Model for action of histone modifications at replication origins.
Replication factors have differential affinity for chromatin regions. Low affinity occurs at regions with low levels of histone acetylation and absence of K36me (silenced chromatin, left) and high levels of K36me3 (transcribed chromatin, right). High levels of K36me1 and histone acetylation cause high affinity for replication factors (center).
Similar articles
- Histone H3 lysine 36 methylation antagonizes silencing in Saccharomyces cerevisiae independently of the Rpd3S histone deacetylase complex.
Tompa R, Madhani HD. Tompa R, et al. Genetics. 2007 Feb;175(2):585-93. doi: 10.1534/genetics.106.067751. Epub 2006 Dec 18. Genetics. 2007. PMID: 17179083 Free PMC article. - Origin association of Sld3, Sld7, and Cdc45 proteins is a key step for determination of origin-firing timing.
Tanaka S, Nakato R, Katou Y, Shirahige K, Araki H. Tanaka S, et al. Curr Biol. 2011 Dec 20;21(24):2055-63. doi: 10.1016/j.cub.2011.11.038. Epub 2011 Dec 8. Curr Biol. 2011. PMID: 22169533 - Methylation of histone H3 at lysine 37 by Set1 and Set2 prevents spurious DNA replication.
Santos-Rosa H, Millán-Zambrano G, Han N, Leonardi T, Klimontova M, Nasiscionyte S, Pandolfini L, Tzelepis K, Bartke T, Kouzarides T. Santos-Rosa H, et al. Mol Cell. 2021 Jul 1;81(13):2793-2807.e8. doi: 10.1016/j.molcel.2021.04.021. Epub 2021 May 11. Mol Cell. 2021. PMID: 33979575 Free PMC article. - Behavior of replication origins in Eukaryota - spatio-temporal dynamics of licensing and firing.
Musiałek MW, Rybaczek D. Musiałek MW, et al. Cell Cycle. 2015;14(14):2251-64. doi: 10.1080/15384101.2015.1056421. Epub 2015 Jun 1. Cell Cycle. 2015. PMID: 26030591 Free PMC article. Review. - A site to remember: H3K36 methylation a mark for histone deacetylation.
Lee JS, Shilatifard A. Lee JS, et al. Mutat Res. 2007 May 1;618(1-2):130-4. doi: 10.1016/j.mrfmmm.2006.08.014. Epub 2007 Jan 21. Mutat Res. 2007. PMID: 17346757 Review.
Cited by
- Chromatin-dependent and -independent regulation of DNA replication origin activation in budding yeast.
Lõoke M, Kristjuhan K, Värv S, Kristjuhan A. Lõoke M, et al. EMBO Rep. 2013 Feb;14(2):191-8. doi: 10.1038/embor.2012.196. Epub 2012 Dec 7. EMBO Rep. 2013. PMID: 23222539 Free PMC article. - Combinatorial modeling of chromatin features quantitatively predicts DNA replication timing in Drosophila.
Comoglio F, Paro R. Comoglio F, et al. PLoS Comput Biol. 2014 Jan;10(1):e1003419. doi: 10.1371/journal.pcbi.1003419. Epub 2014 Jan 23. PLoS Comput Biol. 2014. PMID: 24465194 Free PMC article. - Noncoding transcription influences the replication initiation program through chromatin regulation.
Soudet J, Gill JK, Stutz F. Soudet J, et al. Genome Res. 2018 Dec;28(12):1882-1893. doi: 10.1101/gr.239582.118. Epub 2018 Nov 6. Genome Res. 2018. PMID: 30401734 Free PMC article. - Where and when to start: Regulating DNA replication origin activity in eukaryotic genomes.
Lee CSK, Weiβ M, Hamperl S. Lee CSK, et al. Nucleus. 2023 Dec;14(1):2229642. doi: 10.1080/19491034.2023.2229642. Nucleus. 2023. PMID: 37469113 Free PMC article. Review. - A PWWP domain-containing protein targets the NuA3 acetyltransferase complex via histone H3 lysine 36 trimethylation to coordinate transcriptional elongation at coding regions.
Gilbert TM, McDaniel SL, Byrum SD, Cades JA, Dancy BC, Wade H, Tackett AJ, Strahl BD, Taverna SD. Gilbert TM, et al. Mol Cell Proteomics. 2014 Nov;13(11):2883-95. doi: 10.1074/mcp.M114.038224. Epub 2014 Aug 6. Mol Cell Proteomics. 2014. PMID: 25104842 Free PMC article.
References
- Diffley JF. Regulation of early events in chromosome replication. Curr Biol. 2004;14:R778–786. - PubMed
- Vogelauer M, Rubbi L, Lucas I, Brewer BJ, Grunstein M. Histone acetylation regulates the time of replication origin firing. Mol Cell. 2002;10:1223–1233. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials