The Ca(2+) channel TRPML3 regulates membrane trafficking and autophagy - PubMed (original) (raw)
The Ca(2+) channel TRPML3 regulates membrane trafficking and autophagy
Hyun Jin Kim et al. Traffic. 2009 Aug.
Abstract
TRPML3 is an inward rectifying Ca(2+) channel that is regulated by extracytosolic H(+). Although gain-of-function mutation in TRPML3 causes the varitint-waddler phenotype, the role of TRPML3 in cellular physiology is not known. In this study, we report that TRPML3 is a prominent regulator of endocytosis, membrane trafficking and autophagy. Gradient fractionation and confocal localization reveal that TRPML3 is expressed in the plasma membrane and multiple intracellular compartments. However, expression of TRPML3 is dynamic, with accumulation of TRPML3 in the plasma membrane upon inhibition of endocytosis, and recruitment of TRPML3 to autophagosomes upon induction of autophagy. Accordingly, overexpression of TRPML3 leads to reduced constitutive and regulated endocytosis, increased autophagy and marked exacerbation of autophagy evoked by various cell stressors with nearly complete recruitment of TRPML3 into the autophagosomes. Importantly, both knockdown of TRPML3 by siRNA and expression of the channel-dead dominant negative TRPML3(D458K) have a reciprocal effect, reducing endocytosis and autophagy. These findings reveal a prominent role for TRPML3 in regulating endocytosis, membrane trafficking and autophagy, perhaps by controlling the Ca(2+) in the vicinity of cellular organelles that is necessary to regulate these cellular events.
Conflict of interest statement
All authors declare that they do not have any conflict of interest to disclose.
Figures
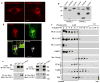
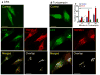
Fig. 1. Expressed TRPML3 is expressed in multiple intracellular compartments
Panel (a): mCherry-TRPML3 was expressed in human skin fibroblasts obtained from TRPML1+/- relative (left image) and TRPML1-/- patient with MLIV (10) (right image) showing that the vesicular expression pattern of TRPML3 is independent of TRPML1. Bars in these and all images donates 10 μm. Panel (b): GFP-TRPML1 and mCherry-TRPML3 were expressed in TRPML1-/- cells and the overlap in expression was determined using imageJ. Panel (c) shows minimal co-IP of HA-TRPML1 and Myc-TRPML3 when co-expressed in HEK cells. The upper blots are the lysates and the lower blots are the co-IP with the indicated antibodies. Panel (d) shows the sensitivity of TRPML3 and TRPML1 to PNGase F and Endo H, and that TRPML3 is not cleaved. Panel (e) show expression of TRPML3 in different subcellular fractions separated on a percoll gradient and expression of TRPML1 only in the heavy fraction. Here and in all blots (ML1) refer to TRPML1 and (ML3) to TRPML3.
Fig. 2. Native TRPML3 is expressed in multiple cellular compartments
In panel (a) HeLa or HEK cells were untreated (Extract), were treated with two different TRPML3 siRNA (si1ML3 and si2ML3), or were transfected with human (hML3) of mouse (mML3) TRPML3 cDNA constructs and were used to probe expression of TRPML3 by western blots with the ProteinTec antibodies (left blot) or the αC2 antibodies (right blot). The arrowheads show that both antibodies recognize the native TRPML3. In panel (b), fractions of untransfected HeLa cells were collected from a percoll gradient as in Fig. 1 and were probed for expression of native TRPML3 (ML3), TRPML1 (ML1), LAMP1 and Transferrin receptor (TrnfR).
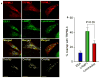
Fig. 3. Subcellular localization of TRPML3
Panel (a): HeLa cells were transfected with mCherry-TRPML3 and either fixed and stained for EEA1 (left images) or LAMP1 (middle images), or live cells were incubated with 10 nM lysotracker green for 10 min (right images) and imaged by confocal microscopy. Panel (b): Images similar to those in panel (a) from at least 10 cells in three different experiments were used to determine the staining overlap with ImageJ. The results are given as mean±s.e.m.
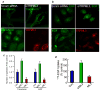
Fig. 4. Overexpression of TRPML3 inhibits transferrin and EGF endocytosis
The blot in panel (a) shows the effect of treating HeLa cells for 48 hrs with TRPML3 siRNA (siTRPML3) on TRPML3 mRNA level. Panels (a, b): The upper images show the effect of siTRPML3 and the lower images the effect of expressing mCherry-TRPML3 on 10 min uptake of transferrin (a) and EGF (b). In panel (c), ImageJ was used to count the number of puncta in 17-19 cells from 4-5 experiments and the fold change is plotted as mean±s.e.m at the indicated conditions. Panel (d) shows the 125I-EGF uptake into cells transfected with TRPML3 or treated with siTRPML3. The results are mean±s.e.m of three experiments.
Fig. 5. TRPML3 regulates endocytosis and membrane trafficking
Panel (a): degradation of EGFR was measured by incubating control HeLa cells and HeLa cells transfected with TRPML3 with 100 ng/ml EGF for the indicated times and analysis of EGFRs by western blot. Tubulin levels were used to control for loading. The 10 min endocytosis averages are the mean±s.e.m. and the effect of TRPML3 is different from control at the p=0.041 level. Panel (b): degradation of EGFRs was measured by incubating control cells and cells treated with TRPML3 siRNA with 100 ng/ml EGF for the indicated times. The 10 min endocytosis averages are the mean±s.e.m. and the effect of TRPML3 siRNA is different from control at the p=0.043 level. Panel (c): degradation of EGFRs was measured in cells transfected with empty vector (control) or with TRPML3(D458,459K) and stimulated with 100 ng/ml EGF for 1 or 5 min. This is one of two experiments with similar results showing the marked increase in EGFR degradation by the DN TRPML3 mutant. Panet (d): degradation of EGFR was measured in control or TRPML1-/- fibroblasts stimulated with 100 ng/ml EGF for the indicated times. This is one of three similar experiments. Panel (e): HeLa cell treated with the two TRPML3 siRNA or transfected with empty vector, TRPML3 or TRPML3(D458K) were used to determine the effect of the various treatments on total and surface expression of EGFR. Total EGFRs (left columns) were determined by densitometry and intensities were first normalized relative to tubulin or actin that were used as loading controls and were then calculated as fold change relative to cells transfected with empty vector. Surface expression (right columns) was normalized relative to the calculated total EGFRs and were then calculated as fold change relative to cells transfected with empty vector. The columns show the m±s.e.m. of 4 experiments.
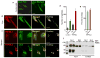
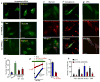
Fig. 6. Dynamic subcellular localization of TRPML3
In panel (a) HeLa cells were transfected with TRPML3 and were kept in fed media or starved for 2 hrs in media without energy substrates. In panel (b) the cells were co-transfected with eGFP-LC3 and mCherry-TRPML3 and maintained in fed media. Panel (c) shows two examples of cells co-transfected with eGFP-LC3 and mCherry-TRPML3 and starved for 2 hrs. The upper images are of cells in which the autophagosomes are not coalesced into large macrophagosomes and the autophagosomes can be resolved and counted. The lower images show the coalesced autophagosomes. Only cells similar to the upper cells in (c) were used for autophagosomes counting, while the two cell types were used to calculate overlap. The number of puncta was counted (d) and the overlap between TRPML3 and LC3 was determined (e) with imageJ and are given as mean±s.e.m of at least 8 cells. Panel (f) shows total (input) and surface expression of TRPML1 and TRPML3 in control cells and cells transfected with the DN dynamin (K44A). Surface expression was determined by biotinylation.
Fig. 7. TRPML3 exacerbates cell-stress induced autophagy
HeLa cells transfected with eGFP-LC3 (the upper image in a, b) or with eGFP-LC3 and mCherry-TRPML3 were treated with 25 μM CPA for 3 hrs or with 3 μM tunicamycin for 1 hr and used to determine the formation of macro-autophagosomes and recruitment of mCherry-TRPML3 into the macro-autophagosomes. Panel (c) shows the time course of autophagy induced by 3 μM tunicamycin in cells transfected with empty vector (blue), TRPML3 (red) or TRPML3(D458K) (green) and left untreated or were treated with tunicamycin for 10 or 30 min. The results are the mea±s.e.m. from at least 6 cells at each time. The dashed line show the change in the number of autophagosomes in the first 10 min of incubation with tunicamycin.
Fig. 8. Inhibition of TRPML3 channel activity inhibits cell-stress induced autophagy
HeLa cells were treated with scrambled (a) or two different siTRPML3 (b) and were incubated in feeding media or were starved for 2 hrs. The average number of puncta is shown in (c) as the mean±s.e.m of at least 7 cells from 2 experiments. Panel (d) shows the Na+ current measured in cells expressing wild-type GFP-TRPML3, GFP-TRPML3(D458K) and TRPML3+ TRPML3(D458K). TRPML3 was activated by exposing the cells to Na+-free medium and then the Na+ current was measured by re-addition of Na+ to the medium. The procedure for current measurement is detailed in Methods and the properties of the TRPML3 current can be found in (8). Note the nearly complete inhibition of TRPML3 current by the DN TRPML3(D458K). In (e-h) the cells were transfected with eGFP-LC3 and the DN mCherry-TRPML3(D458K) and were starved for 2 hrs (e), treated with tunicamycin (f) or CPA (g). The mean±s.e.m of at least 9 cells from 3 experiments is given in (h).
Similar articles
- Palmitoylation controls trafficking of the intracellular Ca2+ channel MCOLN3/TRPML3 to regulate autophagy.
Kim SW, Kim DH, Park KS, Kim MK, Park YM, Muallem S, So I, Kim HJ. Kim SW, et al. Autophagy. 2019 Feb;15(2):327-340. doi: 10.1080/15548627.2018.1518671. Epub 2018 Sep 14. Autophagy. 2019. PMID: 30215288 Free PMC article. - Gain-of-function mutation in TRPML3 causes the mouse Varitint-Waddler phenotype.
Kim HJ, Li Q, Tjon-Kon-Sang S, So I, Kiselyov K, Muallem S. Kim HJ, et al. J Biol Chem. 2007 Dec 14;282(50):36138-42. doi: 10.1074/jbc.C700190200. Epub 2007 Oct 25. J Biol Chem. 2007. PMID: 17962195 - The Ca2+ channel TRPML3 specifically interacts with the mammalian ATG8 homologue GATE16 to regulate autophagy.
Choi S, Kim HJ. Choi S, et al. Biochem Biophys Res Commun. 2014 Jan 3;443(1):56-61. doi: 10.1016/j.bbrc.2013.11.044. Epub 2013 Nov 20. Biochem Biophys Res Commun. 2014. PMID: 24269818 - TRPML3 and hearing loss in the varitint-waddler mouse.
Atiba-Davies M, Noben-Trauth K. Atiba-Davies M, et al. Biochim Biophys Acta. 2007 Aug;1772(8):1028-31. doi: 10.1016/j.bbadis.2007.01.007. Epub 2007 Jan 23. Biochim Biophys Acta. 2007. PMID: 17329082 Review. - The varitint-waddler mouse phenotypes and the TRPML3 ion channel mutation: cause and consequence.
Cuajungco MP, Samie MA. Cuajungco MP, et al. Pflugers Arch. 2008 Nov;457(2):463-73. doi: 10.1007/s00424-008-0523-4. Epub 2008 May 27. Pflugers Arch. 2008. PMID: 18504603 Review.
Cited by
- The Interplay between Ca2+ Signaling Pathways and Neurodegeneration.
Ureshino RP, Erustes AG, Bassani TB, Wachilewski P, Guarache GC, Nascimento AC, Costa AJ, Smaili SS, Pereira GJDS. Ureshino RP, et al. Int J Mol Sci. 2019 Nov 28;20(23):6004. doi: 10.3390/ijms20236004. Int J Mol Sci. 2019. PMID: 31795242 Free PMC article. Review. - Mucolipin co-deficiency causes accelerated endolysosomal vacuolation of enterocytes and failure-to-thrive from birth to weaning.
Remis NN, Wiwatpanit T, Castiglioni AJ, Flores EN, Cantú JA, García-Añoveros J. Remis NN, et al. PLoS Genet. 2014 Dec 18;10(12):e1004833. doi: 10.1371/journal.pgen.1004833. eCollection 2014 Dec. PLoS Genet. 2014. PMID: 25521295 Free PMC article. - NAADP-mediated Ca2+ signaling promotes autophagy and protects against LPS-induced liver injury.
Rah SY, Lee YH, Kim UH. Rah SY, et al. FASEB J. 2017 Jul;31(7):3126-3137. doi: 10.1096/fj.201601290R. Epub 2017 Apr 6. FASEB J. 2017. PMID: 28386045 Free PMC article. - TRPML1 links lysosomal calcium to autophagosome biogenesis through the activation of the CaMKKβ/VPS34 pathway.
Scotto Rosato A, Montefusco S, Soldati C, Di Paola S, Capuozzo A, Monfregola J, Polishchuk E, Amabile A, Grimm C, Lombardo A, De Matteis MA, Ballabio A, Medina DL. Scotto Rosato A, et al. Nat Commun. 2019 Dec 10;10(1):5630. doi: 10.1038/s41467-019-13572-w. Nat Commun. 2019. PMID: 31822666 Free PMC article. - Defective autophagosome trafficking contributes to impaired autophagic flux in coronary arterial myocytes lacking CD38 gene.
Zhang Y, Xu M, Xia M, Li X, Boini KM, Wang M, Gulbins E, Ratz PH, Li PL. Zhang Y, et al. Cardiovasc Res. 2014 Apr 1;102(1):68-78. doi: 10.1093/cvr/cvu011. Epub 2014 Jan 20. Cardiovasc Res. 2014. PMID: 24445604 Free PMC article.
References
- Nilius B, Owsianik G, Voets T, Peters JA. Transient receptor potential cation channels in disease. Physiol Rev. 2007;87(1):165–217. - PubMed
- Bargal R, Avidan N, Ben-Asher E, Olender Z, Zeigler M, Frumkin A, Raas-Rothschild A, Glusman G, Lancet D, Bach G. Identification of the gene causing mucolipidosis type IV. Nature genetics. 2000;26(1):118–123. - PubMed
- Kim HJ, Li Q, Tjon-Kon-Sang S, So I, Kiselyov K, Muallem S. Gain-of-function mutation in TRPML3 causes the mouse varitint-waddler phenotype. J Biol Chem. 2007 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 DE012309-14/DE/NIDCR NIH HHS/United States
- R01 DE012309-13/DE/NIDCR NIH HHS/United States
- R01 DE012309/DE/NIDCR NIH HHS/United States
- R01 DK038938-24S1/DK/NIDDK NIH HHS/United States
- DE12309/DE/NIDCR NIH HHS/United States
- R01 DK038938/DK/NIDDK NIH HHS/United States
- DK38938/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous