H3K4me3 stimulates the V(D)J RAG complex for both nicking and hairpinning in trans in addition to tethering in cis: implications for translocations - PubMed (original) (raw)
H3K4me3 stimulates the V(D)J RAG complex for both nicking and hairpinning in trans in addition to tethering in cis: implications for translocations
Noriko Shimazaki et al. Mol Cell. 2009.
Abstract
The PHD finger of the RAG2 polypeptide of the RAG1/RAG2 complex binds to the histone H3 modification, trimethylated lysine 4 (H3K4me3), and in some manner increases V(D)J recombination. In the absence of biochemical studies of H3K4me3 on purified RAG enzyme activity, the precise role of H3K4me3 remains unclear. Here, we find that H3K4me3 stimulates purified RAG enzymatic activity at both the nicking (2- to 5-fold) and hairpinning (3- to 11-fold) steps of V(D)J recombination. Remarkably, this stimulation can be achieved with free H3K4me3 peptide (in trans), indicating that H3K4me3 functions via two distinct mechanisms. It not only tethers the RAG enzyme complex to a region of DNA, but it also induces a substantial increase in the catalytic turnover number (k(cat)) of the RAG complex. The H3K4me3 catalytic stimulation applies to suboptimal cryptic RSS sites located at H3K4me3 peaks that are critical in the inception of human T cell acute lymphoblastic lymphomas.
Figures
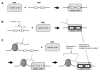
Figure 1. H3K4me3 Tethering Action versus Catalytic Stimulation of the RAG Complex in Cis or Trans
These diagrams depict interactions of the RAG complex, an RSS, and the H3K4me3 peptide in ways that are known (A) or which are tested and supported by experiments here (B and C). (A) The RAG complex binds to the RSS (and can subsequently catalyze nicking and hairpin formation). Within the RSS, the heptamer is designated as `7', and the nonamer is designated as `9'. The catalytic center refers to the portion that binds to the RSS and carries out the catalysis. (B) The RAG complex acquires improved catalytic activity through its binding to free H3K4me3 peptide in trans. Note the difference in shape of the RAG active site, signifying that a conformational change is induced by the H3K4me3. (C) H3K4me3 bound to the RSS substrate DNA fragment can improve RAG action in two ways, called mechanisms 1 and 2. In mechanism 1, the H3K4me3 provides an independent tethering site for the RAGs (in addition to the RAG binding site for DNA). In mechanism 2, the H3K4me3 induces a conformational change in the RAG catalytic center that improves its catalytic activity.
Figure 2. RSS-Bound H3K4me3 Can Stimulate Cleavage and Binding of the RAG Complex to the Substrate
(A) Representation of H3K4me3-bound RSS oligonucleaotide substrate. Biotin tagged histone H3 peptide and biotin tagged 12- or 23-RSS are tethered together through their binding to a streptavidin (SA) tetramer. Both the top and bottom strands of 12-RSS are labeled with radioisotope as indicated by asterisks. (B) EMSA of H3K4me3 bound RSS oligonucleotide substrate preparation. End-labeled 12-RSS tagged with a biotin molecule on the 3' of the bottom strand (lane 1) was incubated with 1.5-fold more SA relative to the RSS DNA (lane 2). Then 2-fold more unmodified H3 or H3K4me3 peptide relative to SA was added to the RSS:SA complex (lane 3, 4). DNA complex was fractionated by 6% non-denaturing PAGE. The inferred compositions of various species are noted at the side of the panel. SA and H3 peptide bound unlabeled 23-RSS tagged was prepared in the same manner. (C) RSS-bound H3K4me3-activated RAG cleavage. The coupled cleavage assay was done with the substrate that was prepared in (B). End-labeled 12-RSS and 23-RSS bound with SA (lanes 5–8), and unmodified H3 (lanes 9–12), or H3K4me3 peptide (lanes 13–16) was incubated with c/c, f/c, c/f RAGs and HMGB1 protein for 1 hr at 37 °C. The products were analyzed on 10 % denaturing PAGE and followed by autoradiography. The positions of substrate (S), nick (N), signal end and hairpin (HP coding end) products are indicated in the right margin. Since both the top and bottom strand are end-labeled, both signal and coding end products could be visualized here.
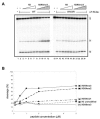
Figure 3. Free H3K4me3 Peptide Stimulates the Cleavage Activity of a RAG Complex That Includes Full-Length
A radiolabeled 12-RSS oligonucleotide substrate and unlabeled 23-RSS were incubated with c/c (lanes 2–10), f/c (lanes 11–19), or c/f (lanes 20–28) RAG proteins (0.1 μM) in the absence or presence of increasing amounts of unmodified H3 or H3K4me3 (0.1, 0.5, 2, 5 μM) peptide for 1 hr at 37 °C. The products were analyzed on 10% denaturing PAGE, followed by autoradiography here and in subsequent figures. The positions of substrate (S), nicked (N), and hairpin (H) products are indicated in the right margin here and in subsequent figures.
Figure 4. A PHD Finger Point Mutant of RAG2 Can No Longer be Stimulated by H3K4me3 and the H3K4Me3 Effect is Specific
(A) The W453R point mutant was previously shown not to bind the PHD finger. Here the RAG catalytic activity is tested for c/f RAG complexes that do or do not have the RAG2 PHD finger point mutation at 453. A radiolabeled 12-RSS oligonucleotide substrate and unlabeled 23-RSS were incubated with 0.1 μM WT of c/f RAG proteins (lanes 2–12) or RAG2 W453R mutant c/f RAG proteins (lanes 13–23) in the absence or presence of increasing amounts (0.1, 0.5, 1, 2, 5 μM) of unmodified H3 or H3K4me3 peptide for 1 hr at 37 °C. The stimulation is abolished by the W453R mutation. (B) The degree of methylation at H3K4 determines the strength of stimulation of RAG nicking and hairpin catalytic activity in a manner that is very similar to the known effects for binding to the PHD finger (Ramón-Maiques et al., 2007). A radiolabeled 12-RSS oligonucleotide substrate and unlabeled 23-RSS were incubated with 0.1 μM c/f RAG proteins in the absence or presence of increasing amounts (0.1, 0.5, 1, 2, 5 μM) of unmodified H3 (indicated as squares), H3K4me1 (triangles), H3K4me2 (diamond), H3K4me3 (filled circles) or H3K9me3 (open circles) peptide for 1 hr at 37°C. The plotted results are shown.
Figure 5. Free H3K4me3 Peptide Stimulates the Nicking by the RAG Complex
(A) A time course of nicking for a radiolabeled 12-RSS oligonucleotide substrate by c/f RAG proteins (0.1 μM) in the absence or presence of unmodified H3 or H3K4me3 (1 μM) peptide is shown in (A). (B) The amount of nicked products obtained in panel (A) was plotted in (B) after phosphorimager quantitation. The amount of nicked products without peptide are shown as triangles, with unmodified H3 peptide as squares, and H3K4me3 peptide as circles here and in (D) below. (C) A time course of nicking for a radiolabeled 23-RSS oligonucleotide substrate by c/f RAG proteins (0.1 μM) in the absence or presence of unmodified H3 or H3K4me3 (1 μM) peptide is shown in (C). The products were analyzed and designated as in (A). Note that above the hairpin position there is a very small amount of aberrant nicking. (D) Nicked products obtained in panel (C) were quantified using phosphorimaging and plotted.
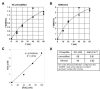
Figure 6. H3K4me3 Increases the Catalytic Turnover Number of the RAG Complex
(A and B) Initial rate kinetics were done for the conversion of 12-RSS to nicked product by the c/f RAG complex in the presence of unmodified H3K4 peptide (A) or in the presence of H3K4me3 peptide (B). This was done at various concentrations of substrate. (C) Determining of the active c/f RAG concentration by burst kinetics. End-labeled 12-RSS was incubated with 40, 70, 100 nM c/f RAG proteins for a 20 min time course and the active fraction was determined as described previously (Yu and Lieber, 2000). Burst kinetics permit determination that 3.4% of the c/f RAG complexes are catalytically active. (D) Kinetic constants was determined based on the best-fit curves (DeltaGraph5.6.4) of the initial rate versus substrate concentration studies in (A) and (B) and the fraction and active c/f RAG proteins.
Figure 7. Free H3K4me3 Peptide Stimulates Hairpin Formation by the RAG Complex
(A) A time course of hairpin formation for a radiolabeled pre-nicked 12-RSS and unlabeled pre-nicked 23-RSS oligonucleotide substrate by c/f RAG proteins (0.1 μM) in the absence or presence of unmodified H3 or H3K4me3 (1 μM) peptide is shown in (A). The products were analyzed on 10 % denaturing PAGE and followed by autoradiography. The positions of prenicked substrate (S, (N)), and hairpin (H) products are indicated in the right margin. (B) The amount of hairpin products obtained in panel (A) was plotted in (B) after phosphorimager quantitation. The amount of hairpin products without peptide are shown as triangles, unmodified H3 peptide as squares, and H3K4me3 peptide as circles.
Comment in
- A histone code for regulating V(D)J recombination.
Schlissel MS, Schulz D, Vettermann C. Schlissel MS, et al. Mol Cell. 2009 Jun 26;34(6):639-40. doi: 10.1016/j.molcel.2009.06.004. Mol Cell. 2009. PMID: 19560416
Similar articles
- Autoinhibition of DNA cleavage mediated by RAG1 and RAG2 is overcome by an epigenetic signal in V(D)J recombination.
Grundy GJ, Yang W, Gellert M. Grundy GJ, et al. Proc Natl Acad Sci U S A. 2010 Dec 28;107(52):22487-92. doi: 10.1073/pnas.1014958107. Epub 2010 Dec 13. Proc Natl Acad Sci U S A. 2010. PMID: 21149691 Free PMC article. - RAG2 PHD finger couples histone H3 lysine 4 trimethylation with V(D)J recombination.
Matthews AG, Kuo AJ, Ramón-Maiques S, Han S, Champagne KS, Ivanov D, Gallardo M, Carney D, Cheung P, Ciccone DN, Walter KL, Utz PJ, Shi Y, Kutateladze TG, Yang W, Gozani O, Oettinger MA. Matthews AG, et al. Nature. 2007 Dec 13;450(7172):1106-10. doi: 10.1038/nature06431. Epub 2007 Nov 21. Nature. 2007. PMID: 18033247 Free PMC article. - Binding and allosteric transmission of histone H3 Lys-4 trimethylation to the recombinase RAG-1 are separable functions of the RAG-2 plant homeodomain finger.
May MR, Bettridge JT, Desiderio S. May MR, et al. J Biol Chem. 2020 Jul 3;295(27):9052-9060. doi: 10.1074/jbc.RA120.014382. Epub 2020 May 15. J Biol Chem. 2020. PMID: 32414844 Free PMC article. - Histone methylation and V(D)J recombination.
Shimazaki N, Lieber MR. Shimazaki N, et al. Int J Hematol. 2014 Sep;100(3):230-7. doi: 10.1007/s12185-014-1637-4. Epub 2014 Jul 25. Int J Hematol. 2014. PMID: 25060705 Review. - Structural gymnastics of RAG-mediated DNA cleavage in V(D)J recombination.
Ru H, Zhang P, Wu H. Ru H, et al. Curr Opin Struct Biol. 2018 Dec;53:178-186. doi: 10.1016/j.sbi.2018.11.001. Epub 2018 Nov 23. Curr Opin Struct Biol. 2018. PMID: 30476719 Free PMC article. Review.
Cited by
- Epigenetic regulation of genomic integrity.
Deem AK, Li X, Tyler JK. Deem AK, et al. Chromosoma. 2012 Apr;121(2):131-51. doi: 10.1007/s00412-011-0358-1. Epub 2012 Jan 17. Chromosoma. 2012. PMID: 22249206 Free PMC article. Review. - Histone H3 lysine 4 methylation revisited.
Kusch T. Kusch T. Transcription. 2012 Nov-Dec;3(6):310-4. doi: 10.4161/trns.21911. Epub 2012 Nov 1. Transcription. 2012. PMID: 23117820 Free PMC article. - Both CpG methylation and activation-induced deaminase are required for the fragility of the human bcl-2 major breakpoint region: implications for the timing of the breaks in the t(14;18) translocation.
Cui X, Lu Z, Kurosawa A, Klemm L, Bagshaw AT, Tsai AG, Gemmell N, Müschen M, Adachi N, Hsieh CL, Lieber MR. Cui X, et al. Mol Cell Biol. 2013 Mar;33(5):947-57. doi: 10.1128/MCB.01436-12. Epub 2012 Dec 21. Mol Cell Biol. 2013. PMID: 23263985 Free PMC article. - Epigenetic regulation and T-cell responses in endometriosis - something other than autoimmunity.
Szukiewicz D. Szukiewicz D. Front Immunol. 2022 Jul 22;13:943839. doi: 10.3389/fimmu.2022.943839. eCollection 2022. Front Immunol. 2022. PMID: 35935991 Free PMC article. Review. - Insights into RAG Evolution from the Identification of "Missing Link" Family A RAGL Transposons.
Martin EC, Le Targa L, Tsakou-Ngouafo L, Fan TP, Lin CY, Xiao J, Huang Z, Yuan S, Xu A, Su YH, Petrescu AJ, Pontarotti P, Schatz DG. Martin EC, et al. Mol Biol Evol. 2023 Nov 3;40(11):msad232. doi: 10.1093/molbev/msad232. Mol Biol Evol. 2023. PMID: 37850912 Free PMC article.
References
- Agrawal A, Schatz DG. RAG1 and RAG2 form a stable postcleavage synaptic complex with DNA containing signal ends in V(D)J recombination. Cell. 1997;89:43–53. - PubMed
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, K KZ. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. - PubMed
- Bergeron S, Anderson DK, Swanson PC. RAG and HMGB1 proteins: purification and biochemical analysis of recombination signal complexes. Methods Enzymol. 2006;408:511–528. - PubMed
- Bowen AJ, Corcoran AE. How chromatin remodelling allows shuffling of immunoglobulin heavy chain genes. Mol Biosyst. 2008;4:790–798. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- 5 P30 CA014089/CA/NCI NIH HHS/United States
- R37 CA051105-22/CA/NCI NIH HHS/United States
- R01 GM043236/GM/NIGMS NIH HHS/United States
- R37 CA051105/CA/NCI NIH HHS/United States
- R01 GM043236-17/GM/NIGMS NIH HHS/United States
- R37 CA051105-23/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous