Multiplex padlock targeted sequencing reveals human hypermutable CpG variations - PubMed (original) (raw)
. 2009 Sep;19(9):1606-15.
doi: 10.1101/gr.092213.109. Epub 2009 Jun 12.
Yuan Gao, John Aach, Kun Zhang, Gregory V Kryukov, Bin Xie, Annika Ahlford, Jung-Ki Yoon, Abraham M Rosenbaum, Alexander Wait Zaranek, Emily LeProust, Shamil R Sunyaev, George M Church
Affiliations
- PMID: 19525355
- PMCID: PMC2752131
- DOI: 10.1101/gr.092213.109
Multiplex padlock targeted sequencing reveals human hypermutable CpG variations
Jin Billy Li et al. Genome Res. 2009 Sep.
Abstract
Utilizing the full power of next-generation sequencing often requires the ability to perform large-scale multiplex enrichment of many specific genomic loci in multiple samples. Several technologies have been recently developed but await substantial improvements. We report the 10,000-fold improvement of a previously developed padlock-based approach, and apply the assay to identifying genetic variations in hypermutable CpG regions across human chromosome 21. From approximately 3 million reads derived from a single Illumina Genome Analyzer lane, approximately 94% (approximately 50,500) target sites can be observed with at least one read. The uniformity of coverage was also greatly improved; up to 93% and 57% of all targets fell within a 100- and 10-fold coverage range, respectively. Alleles at >400,000 target base positions were determined across six subjects and examined for single nucleotide polymorphisms (SNPs), and the concordance with independently obtained genotypes was 98.4%-100%. We detected >500 SNPs not currently in dbSNP, 362 of which were in targeted CpG locations. Transitions in CpG sites were at least 13.7 times more abundant than non-CpG transitions. Fractions of polymorphic CpG sites are lower in CpG-rich regions and show higher correlation with human-chimpanzee divergence within CpG versus non-CpG sites. This is consistent with the hypothesis that methylation rate heterogeneity along chromosomes contributes to mutation rate variation in humans. Our success suggests that targeted CpG resequencing is an efficient way to identify common and rare genetic variations. In addition, the significantly improved padlock capture technology can be readily applied to other projects that require multiplex sample preparation.
Figures
Figure 1.
Padlock probe capture of 53,777 CpG sites. (A) The raw probe precursor (150-mer) sample from Agilent (S) was loaded along with a 10-bp ladder (M) on a 6% denaturing PAGE gel. (B) The probe precursors before and after two rounds of PCR amplification were end-sequenced by Illumina Genome Analyzer. (C) The padlock probes were hybridized to the targeted genomic CpG sites with a uniform 40-nt size. To simplify library construction, a target CpG (dot) was located immediately next to the ligation arm of the probe. Enzymatic filling and ligation of the gap (brown) allowed a copy of the target site to form a circle with the padlock probe. The circles were then PCR amplified using the backbone sequences (green) as primers. The common backbone sequence immediately upstream of the ligation arm served as a sequencing primer. (D) Amplification of circles derived from padlock probes. PCR products were loaded on a 6% PAGE gel. The two upper DNA bands had the expected amplicon sizes: 184 bp (subject to gel purification and Illumina sequencing) and 334 bp (if polymerization extended around the circle twice); the lower bands below 50 nt were derived from PCR primers. (Lane M1) 25-bp DNA ladder (Invitrogen); (lanes H.1,H.2) technical replicates of HapMap sample NA10835; (lanes P1,P2,P3,P9,P10) Personal Genomes 1, 2, 3, 9, and 10, respectively; (lane C) no genomic DNA control; (lane M2) low mass DNA ladder (Invitrogen).
Figure 2.
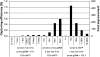
Improvement of padlock capturing efficiency with longer hybridization time (left), more probes (middle), and appropriate dNTP concentration (right). The ratios (10:1, 50:1, 100:1, and 250:1) are molar ratios between each of the padlock probes and genomes. dNTP (1×) is defined as the minimum amount of dNTP needed to capture all genomic copies at each target region. The fold improvement (vertical axis at right) is relative to the reaction with 10:1 probe ratio, hybridized for 15 min at 60°C, and with 100× dNTP. Similar results were observed in independent experiments.
Figure 3.
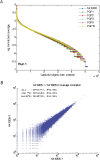
Improved performance of padlock technology. (A) Uniformity of target sites. For each sample, log-normalized coverage levels from sequencing of padlock probe reaction products were computed for each captured target as the log10 of the number of target-mapped, filtered reads divided by the total number of mapped, filtered reads from the reaction. Targets were then ranked for each sample from highest to lowest numbers of mapped, filtered reads and plotted. Except at the extremes, curves exhibit a gradually decreasing slope, indicating that a large number of targets have coverage levels within two orders of magnitude. The plot above depicts sequencing run 1; sequencing run 2 is very similar (Supplemental Fig. 4). For both sequencing runs, overall samples, 54.4%–56.5% of all captured targets had coverage levels within a 10-fold range, and 87.2%–92.7% had coverage within a 100-fold range. (B) Reproducibility of padlock capture. Scatter plot of read coverage of the technical replicate libraries sequenced for NA10835. Pearson correlation coefficients (R) between read counts are provided for all 53,777 target sites (all), all target sites for which one of the replicates has nonzero coverage (one ≠0), and all for which both replicates have nonzero coverage (both ≠0). All Pearson correlation coefficients are >98.1%. The scatter plot is presented on a log–log scale and therefore only contains points corresponding to targets in the “both ≠0” set. The plot above depicts sequencing run 1; sequencing run 2 is very similar (Supplemental Fig. 6). For details on sequencing runs and read mapping and filtering, see text and Supplemental Text.
Figure 4.
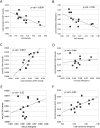
Correlations between polymorphism, interspecies divergence, and CpG content. We analyzed divergence in the chimpanzee lineage after divergence from human using orangutan as an outgroup in order to compensate for bias due to padlock probe design based on the presence of CpGs in the human sequence. SNP densities were calculated as normalized densities per site of a specific type. To calculate the density of CpG SNPs, we divided the total number of the observed CpG polymorphisms in the region by the combined length of all surveyed CpG nucleotides in the region. Correspondingly, to calculate the density of non-CpG SNPs, we divided the total number of observed non-CpG polymorphisms in the region by the combined length of all surveyed non-CpG nucleotides in the region. (A) Correlation between densities of SNPs originated as transitions in CpG sites and fraction of CpGs in the region. (B) Correlation between substitutions due to CpG transitions in the chimpanzee lineage after divergence with humans and fraction of CpGs in the region in the human genome. (C) Correlation between densities of SNPs originated as transitions in CpG sites and non-CpG SNP density. (D) Correlation between densities of SNPs originated as transitions in CpG sites with non-CpG divergence in the chimpanzee lineage after split with humans. (E) Correlation between non-CpG SNP density with non-CpG divergence in the chimpanzee lineage after split with humans. (F) Correlation between densities of SNPs originated as transitions in CpG sites with divergence in the chimpanzee lineage due to CpG transitions.
Similar articles
- Allele-specific methylation is prevalent and is contributed by CpG-SNPs in the human genome.
Shoemaker R, Deng J, Wang W, Zhang K. Shoemaker R, et al. Genome Res. 2010 Jul;20(7):883-9. doi: 10.1101/gr.104695.109. Epub 2010 Apr 23. Genome Res. 2010. PMID: 20418490 Free PMC article. - Sequence context analysis of 8.2 million single nucleotide polymorphisms in the human genome.
Zhao Z, Zhang F. Zhao Z, et al. Gene. 2006 Feb 1;366(2):316-24. doi: 10.1016/j.gene.2005.08.024. Epub 2005 Nov 28. Gene. 2006. PMID: 16314054 - Monitoring methylation changes in cancer.
Beier V, Mund C, Hoheisel JD. Beier V, et al. Adv Biochem Eng Biotechnol. 2007;104:1-11. doi: 10.1007/10_024. Adv Biochem Eng Biotechnol. 2007. PMID: 17290816 Review. - dbSNP in the detail and copy number complexities.
Day IN. Day IN. Hum Mutat. 2010 Jan;31(1):2-4. doi: 10.1002/humu.21149. Hum Mutat. 2010. PMID: 20024941 Review.
Cited by
- Single-cell sequencing-based technologies will revolutionize whole-organism science.
Shapiro E, Biezuner T, Linnarsson S. Shapiro E, et al. Nat Rev Genet. 2013 Sep;14(9):618-30. doi: 10.1038/nrg3542. Epub 2013 Jul 30. Nat Rev Genet. 2013. PMID: 23897237 Review. - Tackling the epigenome: challenges and opportunities for collaboration.
Satterlee JS, Schübeler D, Ng HH. Satterlee JS, et al. Nat Biotechnol. 2010 Oct;28(10):1039-44. doi: 10.1038/nbt1010-1039. Nat Biotechnol. 2010. PMID: 20944594 - Evaluation of molecular inversion probe versus TruSeq® custom methods for targeted next-generation sequencing.
Almomani R, Marchi M, Sopacua M, Lindsey P, Salvi E, Koning B, Santoro S, Magri S, Smeets HJM, Martinelli Boneschi F, Malik RR, Ziegler D, Hoeijmakers JGJ, Bönhof G, Dib-Hajj S, Waxman SG, Merkies ISJ, Lauria G, Faber CG, Gerrits MM; on behalf on the PROPANE Study Group. Almomani R, et al. PLoS One. 2020 Sep 2;15(9):e0238467. doi: 10.1371/journal.pone.0238467. eCollection 2020. PLoS One. 2020. PMID: 32877464 Free PMC article. - microDuMIP: target-enrichment technique for microarray-based duplex molecular inversion probes.
Yoon JK, Ahn J, Kim HS, Han SM, Jang H, Lee MG, Lee JH, Bang D. Yoon JK, et al. Nucleic Acids Res. 2015 Mar 11;43(5):e28. doi: 10.1093/nar/gku1188. Epub 2014 Nov 20. Nucleic Acids Res. 2015. PMID: 25414325 Free PMC article. - RNA-binding proteins in neurodegeneration: Seq and you shall receive.
Nussbacher JK, Batra R, Lagier-Tourenne C, Yeo GW. Nussbacher JK, et al. Trends Neurosci. 2015 Apr;38(4):226-36. doi: 10.1016/j.tins.2015.02.003. Epub 2015 Mar 9. Trends Neurosci. 2015. PMID: 25765321 Free PMC article. Review.
References
- Albert TJ, Molla MN, Muzny DM, Nazareth L, Wheeler D, Song X, Richmond TA, Middle CM, Rodesch MJ, Packard CJ, et al. Direct selection of human genomic loci by microarray hybridization. Nat Methods. 2007;4:903–905. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources