Racial disparity in pathophysiologic pathways of preterm birth based on genetic variants - PubMed (original) (raw)
Racial disparity in pathophysiologic pathways of preterm birth based on genetic variants
Ramkumar Menon et al. Reprod Biol Endocrinol. 2009.
Abstract
Objective: To study pathophysiologic pathways in spontaneous preterm birth and possibly the racial disparity associating with maternal and fetal genetic variations, using bioinformatics tools.
Methods: A large scale candidate gene association study was performed on 1442 SNPs in 130 genes in a case (preterm birth < 36 weeks) control study (term birth > 37 weeks). Both maternal and fetal DNA from Caucasians (172 cases and 198 controls) and 279 African-Americans (82 cases and 197 controls) were used. A single locus association (genotypic) analysis followed by hierarchical clustering was performed, where clustering was based on p values for significant associations within each race. Using Ingenuity Pathway Analysis (IPA) software, known pathophysiologic pathways in both races were determined.
Results: From all SNPs entered into the analysis, the IPA mapped genes to specific disease functions. Gene variants in Caucasians were implicated in disease functions shared with other known disorders; specifically, dermatopathy, inflammation, and hematological disorders. This may reflect abnormal cervical ripening and decidual hemorrhage. In African-Americans inflammatory pathways were the most prevalent. In Caucasians, maternal gene variants showed the most prominent role in disease functions, whereas in African Americans it was fetal variants. The IPA software was used to generate molecular interaction maps that differed between races and also between maternal and fetal genetic variants.
Conclusion: Differences at the genetic level revealed distinct disease functions and operational pathways in African Americans and Caucasians in spontaneous preterm birth. Differences in maternal and fetal contributions in pregnancy outcome are also different between African Americans and Caucasians. These results present a set of explicit testable hypotheses regarding genetic associations with preterm birth in African Americans and Caucasians.
Figures
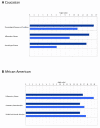
Figure 1
Top disease and disorder functions determined by IPA to be overrepresented by focus genes. Panel A, Caucasian mothers and their fetuses; panel B, African American mothers and their fetuses. Dark blue bars, maternal; light blue, fetal. The p-value for a given diseases and disorder annotation is calculated by the IPA software using Fishers Exact Test taking into account the number of focus genes that participate in that process in relation the total number of genes associated with that process in the IPA knowledgebase.
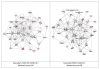
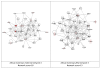
Figure 2
Top ranking gene/protein networks determined by IPA analysis in Caucasian maternal (net work score 34) and fetal (net work score 38). Panel A, maternal networks; panel B, fetal networks. Solid lines show direct interaction (binding/physical contact); dashed line, indirect interaction supported by the literature but possibly involving one or more intermediate molecules that have not been investigated definitively. Molecular interactions involving only binding are connected with a solid line (no arrowhead) since directionality cannot be inferred. Focus genes: pink color, met criteria for case-control comparison for genotype at p ≤ 0.05; red, met criteria for case-control comparison for genotype at p ≤ 0.001; grey, indicating one or more SNP was analyzed in our data set but case-control comparison did not meet p ≤ 0.05; no color – additional interconnected genes generated algorithmically by IPA, i.e., proteins, or complexes, including new potential biomarkers. * indicates that there was more than one SNP probe for this gene tested and the most significant was placed into the analysis.
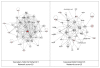
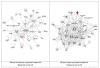
Figure 3
Second ranking gene/protein networks determined by IPA analysis in Caucasian maternal (net work score 32) and fetal (net work score 29). Panel A, maternal networks; panel B, fetal networks. Solid lines show direct interaction (binding/physical contact); dashed line, indirect interaction supported by the literature but possibly involving one or more intermediate molecules that have not been investigated definitively. Molecular interactions involving only binding are connected with a solid line (no arrowhead) since directionality cannot be inferred. Focus genes: pink color, met criteria for case-control comparison for genotype at p ≤ 0.05; red, met criteria for case-control comparison for genotype at p ≤ 0.001; grey, indicating one or more SNP was analyzed in our data set but case-control comparison did not meet p ≤ 0.05; no color – additional interconnected genes generated algorithmically by IPA, i.e., proteins, or complexes, including new potential biomarkers. * indicates that there was more than one SNP probe for this gene tested and the most significant was placed into the analysis.
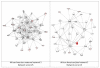
Figure 4
Third ranking Gene/protein networks determined by IPA analysis in Caucasian maternal (net work score 16) and fetal (net work score 13). Panel A, maternal networks; panel B, fetal networks. Solid lines show direct interaction (binding/physical contact); dashed line, indirect interaction supported by the literature but possibly involving one or more intermediate molecules that have not been investigated definitively. Molecular interactions involving only binding are connected with a solid line (no arrowhead) since directionality cannot be inferred. Focus genes: pink color, met criteria for case-control comparison for genotype at p ≤ 0.05; red, met criteria for case-control comparison for genotype at p ≤ 0.001; grey, indicating one or more SNP was analyzed in our data set but case-control comparison did not meet p ≤ 0.05; no color – additional interconnected genes generated algorithmically by IPA, i.e., proteins, or complexes, including new potential biomarkers. * indicates that there was more than one SNP probe for this gene tested and the most significant was placed into the analysis.
Figure 5
Top ranking gene/protein networks determined by IPA analysis in African Americans maternal (net work score 33) and fetal (net work score 40). Panel A, maternal networks; panel B, fetal networks. Solid lines show direct interaction (binding/physical contact); dashed line, indirect interaction supported by the literature but possibly involving one or more intermediate molecules that have not been investigated definitively. Molecular interactions involving only binding are connected with a solid line (no arrowhead) since directionality cannot be inferred. Focus genes: pink color, met criteria for case-control comparison for genotype at p ≤ 0.05; red, met criteria for case-control comparison for genotype at p ≤ 0.001; grey, indicating one or more SNP was analyzed in our data set but case-control comparison did not meet p ≤ 0.05; no color – additional interconnected genes generated algorithmically by IPA, i.e., proteins, or complexes, including new potential biomarkers. * indicates that there was more than one SNP probe for this gene tested and the most significant was placed into the analysis.
Figure 6
Second ranking gene/protein networks determined by IPA analysis in African Americans maternal (net work score 25) and fetal (net work score 15). Panel A, maternal networks; panel B, fetal networks. Solid lines show direct interaction (binding/physical contact); dashed line, indirect interaction supported by the literature but possibly involving one or more intermediate molecules that have not been investigated definitively. Molecular interactions involving only binding are connected with a solid line (no arrowhead) since directionality cannot be inferred. Focus genes: pink color, met criteria for case-control comparison for genotype at p ≤ 0.05; red, met criteria for case-control comparison for genotype at p ≤ 0.001; grey, indicating one or more SNP was analyzed in our data set but case-control comparison did not meet p ≤ 0.05; no color – additional interconnected genes generated algorithmically by IPA, i.e., proteins, or complexes, including new potential biomarkers. * indicates that there was more than one SNP probe for this gene tested and the most significant was placed into the analysis.
Figure 7
Third ranking Gene/protein networks determined by IPA analysis in Caucasian maternal (net work score 18) and fetal (net work score 13). Panel A, maternal networks; panel B, fetal networks. Solid lines show direct interaction (binding/physical contact); dashed line, indirect interaction supported by the literature but possibly involving one or more intermediate molecules that have not been investigated definitively. Molecular interactions involving only binding are connected with a solid line (no arrowhead) since directionality cannot be inferred. Focus genes: pink color, met criteria for case-control comparison for genotype at p ≤ 0.05; red, met criteria for case-control comparison for genotype at p ≤ 0.001; grey, indicating one or more SNP was analyzed in our data set but case-control comparison did not meet p ≤ 0.05; no color – additional interconnected genes generated algorithmically by IPA, i.e., proteins, or complexes, including new potential biomarkers. * indicates that there was more than one SNP probe for this gene tested and the most significant was placed into the analysis.
Figure 8
Fourth ranking Gene/protein networks determined by IPA analysis in Caucasian maternal (net work score 15) and fetal (net work score 13). Panel A, maternal networks; panel B, fetal networks. Solid lines show direct interaction (binding/physical contact); dashed line, indirect interaction supported by the literature but possibly involving one or more intermediate molecules that have not been investigated definitively. Molecular interactions involving only binding are connected with a solid line (no arrowhead) since directionality cannot be inferred. Focus genes: pink color, met criteria for case-control comparison for genotype at p ≤ 0.05; red, met criteria for case-control comparison for genotype at p ≤ 0.001; grey, indicating one or more SNP was analyzed in our data set but case-control comparison did not meet p ≤ 0.05; no color – additional interconnected genes generated algorithmically by IPA, i.e., proteins, or complexes, including new potential biomarkers. * indicates that there was more than one SNP probe for this gene tested and the most significant was placed into the analysis.
Figure 9
Legends used in network diagrams (figures 2–8).
Similar articles
- Dysregulated biomarkers induce distinct pathways in preterm birth.
Brou L, Almli LM, Pearce BD, Bhat G, Drobek CO, Fortunato S, Menon R. Brou L, et al. BJOG. 2012 Mar;119(4):458-73. doi: 10.1111/j.1471-0528.2011.03266.x. BJOG. 2012. PMID: 22324919 - Preterm birth in Caucasians is associated with coagulation and inflammation pathway gene variants.
Velez DR, Fortunato SJ, Thorsen P, Lombardi SJ, Williams SM, Menon R. Velez DR, et al. PLoS One. 2008 Sep 26;3(9):e3283. doi: 10.1371/journal.pone.0003283. PLoS One. 2008. PMID: 18818748 Free PMC article. - Genetic regulation of amniotic fluid TNF-alpha and soluble TNF receptor concentrations affected by race and preterm birth.
Menon R, Velez DR, Morgan N, Lombardi SJ, Fortunato SJ, Williams SM. Menon R, et al. Hum Genet. 2008 Oct;124(3):243-53. doi: 10.1007/s00439-008-0547-z. Epub 2008 Sep 21. Hum Genet. 2008. PMID: 18807256 - Spontaneous preterm birth, a clinical dilemma: etiologic, pathophysiologic and genetic heterogeneities and racial disparity.
Menon R. Menon R. Acta Obstet Gynecol Scand. 2008;87(6):590-600. doi: 10.1080/00016340802005126. Acta Obstet Gynecol Scand. 2008. PMID: 18568457 Review. - Genetic contributions to disparities in preterm birth.
Anum EA, Springel EH, Shriver MD, Strauss JF 3rd. Anum EA, et al. Pediatr Res. 2009 Jan;65(1):1-9. doi: 10.1203/PDR.0b013e31818912e7. Pediatr Res. 2009. PMID: 18787421 Free PMC article. Review.
Cited by
- An overview of racial disparities in preterm birth rates: caused by infection or inflammatory response?
Menon R, Dunlop AL, Kramer MR, Fortunato SJ, Hogue CJ. Menon R, et al. Acta Obstet Gynecol Scand. 2011 Dec;90(12):1325-31. doi: 10.1111/j.1600-0412.2011.01135.x. Epub 2011 May 26. Acta Obstet Gynecol Scand. 2011. PMID: 21615712 Free PMC article. Review. - A spectrum project: preterm birth and small-for-gestational age among infants with birth defects.
Miquel-Verges F, Mosley BS, Block AS, Hobbs CA. Miquel-Verges F, et al. J Perinatol. 2015 Mar;35(3):198-203. doi: 10.1038/jp.2014.180. Epub 2014 Oct 2. J Perinatol. 2015. PMID: 25275696 - Intakes of garlic and dried fruits are associated with lower risk of spontaneous preterm delivery.
Myhre R, Brantsæter AL, Myking S, Eggesbø M, Meltzer HM, Haugen M, Jacobsson B. Myhre R, et al. J Nutr. 2013 Jul;143(7):1100-8. doi: 10.3945/jn.112.173229. Epub 2013 May 22. J Nutr. 2013. PMID: 23700347 Free PMC article. - Interleukin-1 Receptor Antagonist Polymorphism and Birth Timing: Pathway Analysis Among African American Women.
Gillespie SL, Neal JL, Christian LM, Szalacha LA, McCarthy DO, Salsberry PJ. Gillespie SL, et al. Nurs Res. 2017 Mar/Apr;66(2):95-104. doi: 10.1097/NNR.0000000000000200. Nurs Res. 2017. PMID: 28252571 Free PMC article. - Association of IL-4 and IL-4R Polymorphisms with Litter Size Traits in Pigs.
Norseeda W, Liu G, Teltathum T, Supakankul P, Sringarm K, Naraballobh W, Khamlor T, Chomdej S, Nganvongpanit K, Krutmuang P, Mekchay S. Norseeda W, et al. Animals (Basel). 2021 Apr 17;11(4):1154. doi: 10.3390/ani11041154. Animals (Basel). 2021. PMID: 33920608 Free PMC article.
References
- Births Preliminary Data for 2005. National Center for Health Statistics. 2006. - PubMed
- Hogan VK, Richardson JL, Ferre CD, Durant T, Boisseau M. A public health framework for addressing black and white disparities in preterm delivery. J Am Med Womens Assoc. 2001;56:177–180. - PubMed
- Goldenberg RL, Cliver SP, Mulvihill FX, Hickey CA, Hoffman HJ, Klerman LV, Johnson MJ. Medical, psychosocial, and behavioral risk factors do not explain the increased risk for low birth weight among black women. Am J Obstet Gynecol. 1996;175:1317–1324. doi: 10.1016/S0002-9378(96)70048-0. - DOI - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases