CSN-5, a component of the COP9 signalosome complex, regulates the levels of UNC-96 and UNC-98, two components of M-lines in Caenorhabditis elegans muscle - PubMed (original) (raw)
CSN-5, a component of the COP9 signalosome complex, regulates the levels of UNC-96 and UNC-98, two components of M-lines in Caenorhabditis elegans muscle
Rachel K Miller et al. Mol Biol Cell. 2009 Aug.
Abstract
In Caenorhabditis elegans two M-line proteins, UNC-98 and UNC-96, are involved in myofibril assembly and/or maintenance, especially myosin thick filaments. We found that CSN-5, a component of the COP9 signalosome complex, binds to UNC-98 and -96 using the yeast two-hybrid method. These interactions were confirmed by biochemical methods. The CSN-5 protein contains a Mov34 domain. Although one other COP9 signalosome component, CSN-6, also has a Mov34 domain, CSN-6 did not interact with UNC-98 or -96. Anti-CSN-5 antibody colocalized with paramyosin at A-bands in wild type and colocalized with abnormal accumulations of paramyosin found in unc-98, -96, and -15 (encodes paramyosin) mutants. Double knockdown of csn-5 and -6 could slightly suppress the unc-96 mutant phenotype. In the double knockdown of csn-5 and -6, the levels of UNC-98 protein were increased and the levels of UNC-96 protein levels were slightly reduced, suggesting that CSN-5 promotes the degradation of UNC-98 and that CSN-5 stabilizes UNC-96. In unc-15 and unc-96 mutants, CSN-5 protein was reduced, implying the existence of feed back regulation from myofibril proteins to CSN-5 protein levels. Taken together, we found that CSN-5 functions in muscle cells to regulate UNC-98 and -96, two M-line proteins.
Figures
Figure 1.
Yeast two-hybrid assays demonstrate the interaction between specific portions of UNC-96 or -98 with CSN-5. Top right, representation of the UNC-96 and -98 baits used. Left, representation of CSN-5 or -6 preys used. In each case, recognizable protein domains are indicated (C2H2 Zn fingers in UNC-98, Mov34 domains in CSN-5 and -6). Images of growth of three independent colonies of yeast on −Ade plates are shown and summarized as follows: +, growth and −, no growth. Note that interaction of the C-terminal half of UNC-96 or the N-terminal 112 residues of UNC-98 require full-length CSN-5. CSN-6, the other protein of the COP9 complex that contains a Mov34 domain, fails to interact with either UNC-96 or -98.
Figure 2.
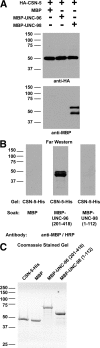
Verification of interaction between CSN-5 and either UNC-96 or -98. (A) Bacterially expressed MBP-UNC-96 (aa 201-418) and MBP-UNC-98 (1-112) interact with yeast expressed HA-CSN-5. Total protein extracts were prepared from yeast expressing HA-CSN-5, incubated with agarose beads coated with antibodies to HA, washed, and then incubated with purified, bacterially expressed MBP, MBP-UNC-96, or MBP-UNC-98, and washed; then the proteins were eluted, and portions of each sample were separated by SDS-PAGE in two gels (10%) and blotted. As shown on top, reaction of one blot with anti-HA shows that in each reaction, HA-CSN-5 is present. As shown on the bottom, reaction with anti-MBP reveals that MBP-UNC-98 was copelleted (top band, full-length product; bottom band, likely degradation product). (B) The C-terminal half of UNC-96 interacts with CSN-5 by far Western assay. SDS-PAGE was used to separate CSN-5-His, and the protein was transferred to a membrane. One blot strip was incubated with MBP, one blot strip was incubated with MBP-UNC-96 (201–418), and a third blot strip was incubated with MBP-UNC-98 (1–112). After washing, each blot strip was incubated with antibodies to MBP coupled to horseradish peroxidase (anti-MBP/HRP), and reactions visualized by ECL. (C) A Coomassie-stained 10% SDS-PAGE shows 2 μg of each bacterially expressed protein used in the far Western or HA pulldown. For each blot or gel, the positions of molecular-weight size markers in kDa are indicated.
Figure 3.
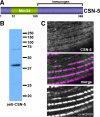
Antibodies raised to CSN-5 detect a polypeptide from worm extracts of expected size and localize to muscle A-bands. (A) Schematic of CSN-5 indicating that a nondomain containing region, residues 166-369, was used as immunogen to generate rabbit polyclonal antibodies. (B) Western blot of whole worm extracts from wild type reacted with affinity-purified antibodies to CSN-5. Major reaction is to an approximately 40-kDa polypeptide, the size expected from the CSN-5 sequence. Molecular-weight standards are shown at the right. (C) Anti-CSN-5 localize to A-bands in body wall muscle. Anti-CSN-5 and anti-α-actinin were coincubated with wild-type C. elegans, and the muscle was imaged by confocal immunofluorescence microscopy. The images show a portion of one body wall muscle cell. α-Actinin is a marker for dense bodies. Bar, 10 μm.
Figure 4.
Immunofluorescence localization of CSN-5 and paramyosin in wild-type and in unc-96 (sf18) and unc-98 (sf19) mutant muscle. In wild-type muscle, CSN-5 localizes to A-bands, colocalizing with paramyosin. In both unc-96 and -98, some CSN-5 is found in A-bands, but much of it is found in needle-like accumulations containing paramyosin at the ends of muscle cells. Bar, 10 μm.
Figure 5.
CSN-5 colocalizes with paramyosin in a paramyosin missense mutant. The paramyosin missense mutant, unc-15 (e1215), has paramyosin paracrystals within its body wall muscle cells. These accumulations of paramyosin also contain CSN-5. Bar, 10 μm.
Figure 6.
Knockdown of csn-5, -6, or both together has no effect on the localization of two thick filament components, MHC A and paramyosin. Wild-type worms were subjected to RNAi by feeding using either the empty vector or with vectors containing the indicated genes. F1 adults were fixed and immunostained with either anti-MHC A or anti-paramyosin. RNAi with the empty vector resulted in worms with the same staining pattern as unfed wild-type worms. Bar, 10 μm.
Figure 7.
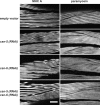
Knockdown of csn-5 and -6 together results in a mild improvement in myofibrillar structure of unc-96 (sf18) animals. Polarized light images of unc-96 (sf18) animals after RNAi by feeding of bacteria harboring either empty vector or with vectors containing the indicated genes. In the worms fed the empty vector note the typical appearance of unc-96 mutant animals with myofilament of reduced organization and birefringent needles at the ends of the muscle cells. RNAi of either csn-5 or -6 alone has no effect on this appearance. However, double RNAi for csn-5 and -6 results in mild improvement, with better organized lattice and fewer or less intense needles (portions of two different animals are shown). Bar, 10 μm.
Figure 8.
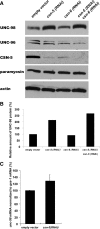
RNAi for csn-5 results in an increase in the levels of UNC-98 protein. (A) Extracts containing equal amounts of total Laemmli-soluble proteins from F1 progeny of animals fed either the RNAi by feeding empty vector or vectors containing csn-5 or -6 or both csn-5 and -6, were separated on a gel, blotted, and reacted with antibodies to the indicated proteins. As expected, the level of CSN-5 is reduced in csn-5 (RNAi) or in csn-5 (RNAi) csn-6 (RNAi). Unexpectedly, CSN-5 is also reduced in csn-6 (RNAi). Note that when csn-5 was knocked down, the level of UNC-98 was increased. The levels of actin and paramyosin served as loading controls. (B) Quantitation of UNC-98 protein levels under different conditions. For each lane, the UNC-98 level was normalized to that of actin. As compared with empty RNAi vector, csn-5 (RNAi) resulted in a 2.1-fold increase, and csn-5 csn-6 double RNAi resulted in a 2.7-fold increase in UNC-98 protein. (C) Quantitative real-time PCR of unc-98 mRNA with and without RNAi for csn-5. mRNA levels of unc-98 were normalized to ges-1. Fold differences in mRNA levels of unc-98 upon csn-5 (RNAi) were calculated relative to the empty vector, which was arbitrarily set at a value of 100. As compared with empty vector, csn-5 (RNAi) resulted in a 1.3-fold increase in unc-98 mRNA levels. The graphs depict means from six experiments, and the error bars depict standard deviations.
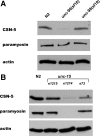
Figure 9.
The effects of mutations in unc-96, -98, and -15 on the levels of CSN-5. Extracts containing equal amounts of total Laemmli-soluble proteins from wild-type (N2), loss-of-function alleles for unc-96 (sf18) and unc-98 (sf19), and the unc-15 null (e1214) and two unc-15 missense mutations (e1215 and e73) were separated on a gel, blotted, and reacted with antibodies to the indicated proteins. (A) The level of CSN-5 protein is diminished in the unc-96 mutant. (Paramyosin, the level of which was shown previously not to be affected in unc-96 or -98 and actin, serve as loading controls.) (B) The level of CSN-5 protein is significantly decreased in the paramyosin null (e1214), and moderately decreased in the two paramyosin missense mutants (especially e73).
Figure 10.
CSN-5 cooperates with UNC-98 and -96 to promote assembly of paramyosin into thick filaments. UNC-98 and -96 are two components of the nematode M-line and interact with each other. Paramyosin, a major component of thick filaments, is likely to exist in two pools: paramyosin incorporated into thick filaments and free paramyosin created by new synthesis or by normal turnover of thick filaments. Previous results supported a model in which UNC-98 and -96 act as chaperones to prevent abnormal aggregation of free paramyosin and thus promote its incorporation into thick filaments. In this study we show that CSN-5, a component of the COP9 signalosome, interacts with both UNC-98 and -96; that CSN-5 promotes the degradation of UNC-98 (arrow), that a combination of CSN-5 and -6 (shown by other studies to interact with each other) stabilize UNC-96 (converging arrows), and that UNC-96 and free paramyosin indirectly or through a positive feedback loop act to increase the levels of CSN-5 (dotted arrows). Overall CSN-5 and -6 act together with UNC-98 and -96 to promote the incorporation of paramyosin into thick filaments.
Similar articles
- Caenorhabditis elegans UNC-96 is a new component of M-lines that interacts with UNC-98 and paramyosin and is required in adult muscle for assembly and/or maintenance of thick filaments.
Mercer KB, Miller RK, Tinley TL, Sheth S, Qadota H, Benian GM. Mercer KB, et al. Mol Biol Cell. 2006 Sep;17(9):3832-47. doi: 10.1091/mbc.e06-02-0144. Epub 2006 Jun 21. Mol Biol Cell. 2006. PMID: 16790495 Free PMC article. - UNC-98 and UNC-96 interact with paramyosin to promote its incorporation into thick filaments of Caenorhabditis elegans.
Miller RK, Qadota H, Mercer KB, Gernert KM, Benian GM. Miller RK, et al. Mol Biol Cell. 2008 Apr;19(4):1529-39. doi: 10.1091/mbc.e07-07-0723. Epub 2008 Feb 6. Mol Biol Cell. 2008. PMID: 18256289 Free PMC article. - The SH3 domain of UNC-89 (obscurin) interacts with paramyosin, a coiled-coil protein, in Caenorhabditis elegans muscle.
Qadota H, Mayans O, Matsunaga Y, McMurry JL, Wilson KJ, Kwon GE, Stanford R, Deehan K, Tinley TL, Ngwa VM, Benian GM. Qadota H, et al. Mol Biol Cell. 2016 May 15;27(10):1606-20. doi: 10.1091/mbc.E15-09-0675. Epub 2016 Mar 23. Mol Biol Cell. 2016. PMID: 27009202 Free PMC article. - The devil is in the details: comparison between COP9 signalosome (CSN) and the LID of the 26S proteasome.
Meister C, Gulko MK, Köhler AM, Braus GH. Meister C, et al. Curr Genet. 2016 Feb;62(1):129-36. doi: 10.1007/s00294-015-0525-7. Curr Genet. 2016. PMID: 26497135 Review. - Mammalian COP9 signalosome.
Kato JY, Yoneda-Kato N. Kato JY, et al. Genes Cells. 2009 Nov;14(11):1209-25. doi: 10.1111/j.1365-2443.2009.01349.x. Epub 2009 Oct 22. Genes Cells. 2009. PMID: 19849719 Review.
Cited by
- Anaplasma phagocytophilum induces actin phosphorylation to selectively regulate gene transcription in Ixodes scapularis ticks.
Sultana H, Neelakanta G, Kantor FS, Malawista SE, Fish D, Montgomery RR, Fikrig E. Sultana H, et al. J Exp Med. 2010 Aug 2;207(8):1727-43. doi: 10.1084/jem.20100276. Epub 2010 Jul 26. J Exp Med. 2010. PMID: 20660616 Free PMC article. - Obscurin and KCTD6 regulate cullin-dependent small ankyrin-1 (sAnk1.5) protein turnover.
Lange S, Perera S, Teh P, Chen J. Lange S, et al. Mol Biol Cell. 2012 Jul;23(13):2490-504. doi: 10.1091/mbc.E12-01-0052. Epub 2012 May 9. Mol Biol Cell. 2012. PMID: 22573887 Free PMC article. - High-resolution x-ray structure and functional analysis of the murine norovirus 1 capsid protein protruding domain.
Taube S, Rubin JR, Katpally U, Smith TJ, Kendall A, Stuckey JA, Wobus CE. Taube S, et al. J Virol. 2010 Jun;84(11):5695-705. doi: 10.1128/JVI.00316-10. Epub 2010 Mar 24. J Virol. 2010. PMID: 20335262 Free PMC article. - Bending amplitude - a new quantitative assay of C. elegans locomotion: identification of phenotypes for mutants in genes encoding muscle focal adhesion components.
Nahabedian JF, Qadota H, Stirman JN, Lu H, Benian GM. Nahabedian JF, et al. Methods. 2012 Jan;56(1):95-102. doi: 10.1016/j.ymeth.2011.11.005. Epub 2011 Nov 22. Methods. 2012. PMID: 22126736 Free PMC article. - Sequence context and crosslinking mechanism affect the efficiency of in vivo capture of a protein-protein interaction.
Lancia JK, Nwokoye A, Dugan A, Joiner C, Pricer R, Mapp AK. Lancia JK, et al. Biopolymers. 2014 Apr;101(4):391-7. doi: 10.1002/bip.22395. Biopolymers. 2014. PMID: 24037947 Free PMC article.
References
- Aamodt E. J., Chung M. A., McGhee J. D. Spatial control of gut-specific gene expression during Caenorhabditis elegans development. Science. 1991;252:579–582. - PubMed
- Cope G. A., Deshaies R. J. COP9 signalosome: multifunctional regulator of SCF and other cullin-based ubiquitin ligases. Cell. 2003;114:663–671. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases