The pluripotency factor Oct4 interacts with Ctcf and also controls X-chromosome pairing and counting - PubMed (original) (raw)
. 2009 Jul 2;460(7251):128-32.
doi: 10.1038/nature08098. Epub 2009 Jun 17.
Affiliations
- PMID: 19536159
- PMCID: PMC3057664
- DOI: 10.1038/nature08098
The pluripotency factor Oct4 interacts with Ctcf and also controls X-chromosome pairing and counting
Mary E Donohoe et al. Nature. 2009.
Abstract
Pluripotency of embryonic stem (ES) cells is controlled by defined transcription factors. During differentiation, mouse ES cells undergo global epigenetic reprogramming, as exemplified by X-chromosome inactivation (XCI) in which one female X chromosome is silenced to achieve gene dosage parity between the sexes. Somatic XCI is regulated by homologous X-chromosome pairing and counting, and by the random choice of future active and inactive X chromosomes. XCI and cell differentiation are tightly coupled, as blocking one process compromises the other and dedifferentiation of somatic cells to induced pluripotent stem cells is accompanied by X chromosome reactivation. Recent evidence suggests coupling of Xist expression to pluripotency factors occurs, but how the two are interconnected remains unknown. Here we show that Oct4 (also known as Pou5f1) lies at the top of the XCI hierarchy, and regulates XCI by triggering X-chromosome pairing and counting. Oct4 directly binds Tsix and Xite, two regulatory noncoding RNA genes of the X-inactivation centre, and also complexes with XCI trans-factors, Ctcf and Yy1 (ref. 17), through protein-protein interactions. Depletion of Oct4 blocks homologous X-chromosome pairing and results in the inactivation of both X chromosomes in female cells. Thus, we have identified the first trans-factor that regulates counting, and ascribed new functions to Oct4 during X-chromosome reprogramming.
Figures
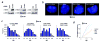
Figure 1. Oct4 and Sox2 bind Tsix and Xite
a, Map of Ctcf, Yy1, Oct4, and Sox2 sites within the 15-kb critical region of the mouse Xic responsible for counting, choice, and pairing. b, The Sox2/Oct4 consensus and representative binding motifs within Tsix, Xite, and ES-specific genes, Fbx15, Utf1, Sox2, Lefty1, Nanog, and Fgf4. c, Coomassie staining of purified recombinant Oct4 and Sox2 proteins (rOct4, rSox2). d, Gel shift of Tsix site E motif using rOct4. Arrow, rOct4-DNA shift. Asterisk, supershifted rOct4-DNA. Comp, cold competitor added at 100x-fold molar excess. e, Gel shift of the Xite motif using rOct4 and rSox2. Arrows, protein-DNA shifts. Asterisks, corresponding supershifts. f, qChIP analysis of Oct4, Sox2, and positive control H3 at designated sites in d0 (day 0) and d4 (day 4) wildtype male and female ES cells. Averages of three independent biological replicates shown with standard error of the mean (S.E.M). Intron 1B results for Sox2 were off-scale, with actual averages +/- S.E.M shown. Statistical significance (P) of each result is calculated against the control IgG ChIP (background) using a paired, one-tailed _t_-test. *, P<0.05.
Figure 2. Oct4-Ctcf and Sox2-Yy1 interactions, and Tsix transcriptional activation
a, Testing mammalian GST fusions of full-length Yy1 and Ctcf or Ctcf domains (amino acids indicated) for interaction with S35-labelled Oct4. Lower panels, α–GST Western control. Arrows, specific protein fusions. b, HEK cells were cotransfected with myc-Ctcf and indicated Flag-tagged Oct4 fragments. Whole cell extracts (WCE) were immunoprecipiated (IP) with α-Flag antibodies prior to Western blotting with α-myc antibodies. Arrow, Ctcf protein. c, Reciprocal co-IP: Left, IP with α-Oct4 or control antibodies to test interaction with endogenous Ctcf; arrow, Ctcf detected by α-Ctcf Western analysis. Right, IP with α-Ctcf or control antibodies to test interaction with endogenous Oct4; arrow, Oct4 detected by α-Oct4 Western. Doublet bands and mobility differences may reflect isoforms due to post-translational modifications. d, Yy1 (S35-labelled) binding to Sox2 but not Oct4 revealed by GST fusion analysis. Bottom, Coomassie staining to reveal GST fusion proteins. e, Domain mapping of Yy1 binding to Sox2. Mobility differences may reflect post-translational modifications. f, Overexpression and co-IP of Flag-Oct4, Myc-Ctcf, and HA-Yy1 in indicated combinations in HEK cells. Two 6-well plates of HEK cells were transfected with 8 μg of tagged constructs and harvested 35 hrs later for coIP analysis. Western blots of the co-IP (top panel) and whole cell extracts (bottom 3 panels) were performed using indicated antibodies. g, Oct4 and Sox2 confer developmental specificity to Xite enhancer. Map of luciferase expression vector fused to the Tsix major promoter (PTsix) and the 1.2 kb enhancer. Mutations of the Oct4/Sox2 motifs in the enhancer compromised PTsix activity in transiently transfected male d0 ES cells. Error bars, 1 SD. h, qRT-PCR for Xite and Tsix RNA after knocking down the indicated factors. Levels are normalized to β-actin levels. Error bars, 1SD. i, Luciferase analysis of cells carrying the wildtype enhancer shows enhanced PTsix activity upon differentiation. Cells were differentiated in duplicate and luciferase levels were normalized to total protein levels.
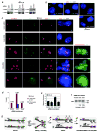
Figure 3. Oct4 knockdown disrupts X-X pairing and results in aberrant Xist expression in female ES cells
a, Western analysis confirms knockdown of indicated proteins in wildtype female ES cells nucleofected with siRNAs on d2 and harvested on d4. b, Representative images of Xic –Xic localization in knockdown cells using a two-probe combination of pSxn (Tsix, green) and pSx9 (Xist, red). To ensure signal specificity, only those foci with overlapping red and green signals are scored. c, Distribution of X-X distances in knockdown cells as indicated. n, nuclei counted. ND= Xic-Xic distance/d, where d = 2 × (nuclear area/π)0.5. The significance of the difference (P) between samples and control siRNA was calculated using the Kolmogorov-Smirnov (KS) test, a non-parametric test to determine whether two data-sets have a similar distribution (SPSS 13.0 software). d, Cumulative frequency curves of the analysis carried out in panel c. ND 0.0-0.2 are shown.
Figure 4. Ectopic Xist expression from both Xs in Oct4-deficient ES cells
a, Western analysis confirms knockdown of indicated proteins in wildtype male ES cells nucleofected on d2 and harvested on d4. b, RNA/DNA FISH analysis for Xist RNA (red) and Xic DNA (green) after indicated knockdowns in d4 male ES cells. c, RNA/DNA FISH shows ectopic Xist upregulation in Oct4- but not Sox2- or control- female knockdown cells on d4. Three representative fields of Oct4-deficient cells are shown. Xist RNA, red. Xic DNA (pSxn probe), green. After RNA/DNA FISH for Xist/Xic, cells were hybridized with a Chr.1 (1C, red) and an Xic (green) probe, revealing that each nucleus with two Xist clusters is diploid. Note that original Xist RNA signals were destroyed in second-round FISH. d, Percentage of cells displaying one versus two Xist RNA clusters on d4. e, Quantitative RT-PCR of Xist RNA in d4 female knockdown cells. f, Allele-specific RT-PCR of Xist in d4 female knockdown cells. Cas, M. castaneus allele. g, Model: Dynamic and multifaceted regulation of XCI by Oct4. +, activating. -, repressive. See text for detailed discussion.
Similar articles
- The BET family member BRD4 interacts with OCT4 and regulates pluripotency gene expression.
Wu T, Pinto HB, Kamikawa YF, Donohoe ME. Wu T, et al. Stem Cell Reports. 2015 Mar 10;4(3):390-403. doi: 10.1016/j.stemcr.2015.01.012. Epub 2015 Feb 12. Stem Cell Reports. 2015. PMID: 25684227 Free PMC article. - Evidence that homologous X-chromosome pairing requires transcription and Ctcf protein.
Xu N, Donohoe ME, Silva SS, Lee JT. Xu N, et al. Nat Genet. 2007 Nov;39(11):1390-6. doi: 10.1038/ng.2007.5. Epub 2007 Oct 21. Nat Genet. 2007. PMID: 17952071 - The X-inactivation trans-activator Rnf12 is negatively regulated by pluripotency factors in embryonic stem cells.
Navarro P, Moffat M, Mullin NP, Chambers I. Navarro P, et al. Hum Genet. 2011 Aug;130(2):255-64. doi: 10.1007/s00439-011-0998-5. Epub 2011 May 5. Hum Genet. 2011. PMID: 21544581 Free PMC article. - The coupling of X-chromosome inactivation to pluripotency.
Deuve JL, Avner P. Deuve JL, et al. Annu Rev Cell Dev Biol. 2011;27:611-29. doi: 10.1146/annurev-cellbio-092910-154020. Epub 2011 Jul 29. Annu Rev Cell Dev Biol. 2011. PMID: 21801017 Review. - Concise review: Pluripotency and the transcriptional inactivation of the female Mammalian X chromosome.
Minkovsky A, Patel S, Plath K. Minkovsky A, et al. Stem Cells. 2012 Jan;30(1):48-54. doi: 10.1002/stem.755. Stem Cells. 2012. PMID: 21997775 Free PMC article. Review.
Cited by
- O-GlcNAc cycling: a link between metabolism and chronic disease.
Bond MR, Hanover JA. Bond MR, et al. Annu Rev Nutr. 2013;33:205-29. doi: 10.1146/annurev-nutr-071812-161240. Epub 2013 Apr 29. Annu Rev Nutr. 2013. PMID: 23642195 Free PMC article. Review. - The Chromatin Signature of Pluripotency: Establishment and Maintenance.
Di Giammartino DC, Apostolou E. Di Giammartino DC, et al. Curr Stem Cell Rep. 2016;2(3):255-262. doi: 10.1007/s40778-016-0055-3. Epub 2016 Jun 27. Curr Stem Cell Rep. 2016. PMID: 27547710 Free PMC article. Review. - Maternal control of early mouse development.
Li L, Zheng P, Dean J. Li L, et al. Development. 2010 Mar;137(6):859-70. doi: 10.1242/dev.039487. Development. 2010. PMID: 20179092 Free PMC article. Review. - Abnormal dosage of ultraconserved elements is highly disfavored in healthy cells but not cancer cells.
McCole RB, Fonseka CY, Koren A, Wu CT. McCole RB, et al. PLoS Genet. 2014 Oct 23;10(10):e1004646. doi: 10.1371/journal.pgen.1004646. eCollection 2014 Oct. PLoS Genet. 2014. PMID: 25340765 Free PMC article. - Single-Cell Landscape of Transcriptional Heterogeneity and Cell Fate Decisions during Mouse Early Gastrulation.
Mohammed H, Hernando-Herraez I, Savino A, Scialdone A, Macaulay I, Mulas C, Chandra T, Voet T, Dean W, Nichols J, Marioni JC, Reik W. Mohammed H, et al. Cell Rep. 2017 Aug 1;20(5):1215-1228. doi: 10.1016/j.celrep.2017.07.009. Cell Rep. 2017. PMID: 28768204 Free PMC article.
References
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126(4):663–676. - PubMed
- Maherali N, et al. Directly Reprogrammed Fibroblasts Show Global Epigenetic Remodeling and Widespread Tissue Contribution. Cell Stem Cell. 2007;1(1):55–70. - PubMed
- Wutz A, Gribnau J. X inactivation Xplained. Curr Opin Genet Dev. 2007;17(5):387–393. - PubMed
- Lucchesi JC, Kelly WG, Panning B. Chromatin remodeling in dosage compensation. Annu Rev Genet. 2005;39:615–651. - PubMed
- Payer B, Lee JT. X Chromosome Dosage Compensation: How Mammals Keep the Balance. Annu Rev Genet. 2008 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- HHMI/Howard Hughes Medical Institute/United States
- R01 GM058839/GM/NIGMS NIH HHS/United States
- R37 GM058839/GM/NIGMS NIH HHS/United States
- R01 GM058839-10/GM/NIGMS NIH HHS/United States
- GM58839/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases