Identification of small gains and losses in single cells after whole genome amplification on tiling oligo arrays - PubMed (original) (raw)
Identification of small gains and losses in single cells after whole genome amplification on tiling oligo arrays
Jochen B Geigl et al. Nucleic Acids Res. 2009 Aug.
Abstract
Clinical DNA is often available in limited quantities requiring whole-genome amplification for subsequent genome-wide assessment of copy-number variation (CNV) by array-CGH. In pre-implantation diagnosis and analysis of micrometastases, even merely single cells are available for analysis. However, procedures allowing high-resolution analyses of CNVs from single cells well below resolution limits of conventional cytogenetics are lacking. Here, we applied amplification products of single cells and of cell pools (5 or 10 cells) from patients with developmental delay, cancer cell lines and polar bodies to various oligo tiling array platforms with a median probe spacing as high as 65 bp. Our high-resolution analyses reveal that the low amounts of template DNA do not result in a completely unbiased whole genome amplification but that stochastic amplification artifacts, which become more obvious on array platforms with tiling path resolution, cause significant noise. We implemented a new evaluation algorithm specifically for the identification of small gains and losses in such very noisy ratio profiles. Our data suggest that when assessed with sufficiently sensitive methods high-resolution oligo-arrays allow a reliable identification of CNVs as small as 500 kb in cell pools (5 or 10 cells), and of 2.6-3.0 Mb in single cells.
Figures
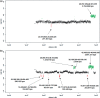
Figure 1.
Ratio profiles of non-amplified DNA of probands P1 (a) and P2 (b) on the NimbleGen Chromosome 22 Tiling array. The calculation of these ratio profiles was based on a classical approach, using a window size of 100 adjacent oligos (corresponding to 6.5 kb) thresholds were simply determined as ±2 times SD. On the NimbleGen arrays losses are illustrated in green above the _X_-axis, whereas gains are shown in red below the _X_-axis. The sizes of observed CNVs are displayed at the respective locations.
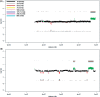
Figure 2.
This figure displays the same ratio profiles as in Figure 1a and b, i.e. the ratio profiles of probands P1 (a) and P2 (b), now calculated with the algorithm described in this manuscript. The center profile is based on calculations with window sizes of 100 adjacent oligos (corresponding to 6.5 kb). A color bar code presents the window size (each in adjacent oligos and the respective physical size) for which calculations have been conducted. In the case of non-amplified DNA we selected very small window sizes, in the other cases with whole genome amplification products the window sizes were larger.
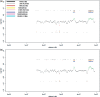
Figure 3.
Cell-pool results obtained for proband P2 on the NimbleGen Chromosome 22 Tiling array. (a) Evaluation of the 10-cell pool on the NimbleGen Chromosome 22 Tiling array. The profile shown in the center was obtained with a window size of 5.000 oligos (corresponding to 325 kb). The two largest CNVs show bar codes from black to cyan, demonstrating that the size of the CNVs is in the range of 1.3 Mb or larger (actual sizes: 3 and 1.2 Mb, respectively; compare Figure 1b). In contrast, the largest duplication has a bar code ranging only from black to blue, showing that the size of this CNV is somewhere between 325 and 650 kb (the actual size is 532 kb, Figure 1b). To the left side of this duplication another region at position 26.5 Mb appears to be potentially duplicated. However, the calls are not uninterrupted from black to blue, as there is no pink bar revealing that this CNV call is likely to be an artifact [compare panel (4) in
Supplementary Figure 2c
]. (b) Hybridization of the 5-cell pool from proband P2 on the NimbleGen Chromosome 22 Tiling array resulted in a CNV recognition pattern similar to that of the 10-cell pool. The algorithm shows the presence of the 255 kb large duplication at position of about 44.7–44.8 (compare Figure 1b), however, the larger 296 and 335 kb duplications were not identified.
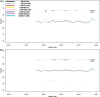
Figure 4.
Chromosome 22 profile for proband P2 obtained with a single cell amplification product on the NimbleGen Chromosome 22 Tiling array. Beside the 3 Mb deletion, the bar code pattern displays a possible presence of two smaller deletions at positions 34 and 38 Mb with sizes between 650 kb and 1.3 Mb. The deletion at position 38 Mb corresponds to the location of the real existing 1.2 Mb deletion. However, the second putative deletion at position 34 Mb is false positive, demonstrating that CNVs with a size of <2 Mb cannot be reliably detected in a single cell. Here the center profile was obtained with a 20.000 oligo sliding window (1.3 Mb).
Figure 5.
Cell-pool results obtained for proband P1 on the NimbleGen Chromosome 22 Tiling array. (a) Hybridization of the 10-cell pool clearly identified the 2.8 Mb-deletion. The algorithm also identified another deletion with a size of about 650 kb at position 21 Mb. This deletion is likely to be an artifact (compare
Supplementary Figure 6b
and details in text). (b) The 5-cell pool of proband P1 also allowed precise identification of the 2.8 Mb-deletion. In addition, at positions 27 and 32 Mb, the algorithm shows the possible presence of two further deletions, each with a size below the 500 kb limit for reliable CNV identification in cell pools. At position 23–24 Mb some bar codes reveal a duplication, which in fact corresponds to the real 272 kb duplication. In both cases the center profile was obtained with a sliding window of 5.000 oligos (325 kb).
Figure 6.
Identification of the 2.8 Mb deletion in a single cell (‘#1’) of proband P1 on the NimbleGen Chromosome 22 Tiling array. The center profile was generated using a 20.000 oligo sliding window (1.3 Mb).
Figure 7.
Ratio profiles of the X-chromosome. (a) Evaluation of the X-chromosome with non-amplified DNA. All X-chromosome landmark regions, i.e. PAR1, PAR2 and the XY-homology region (compare
Supplementary Figure 8a
) are identified. (b) X-chromosome evaluation of the 10-cell pool, which results in a similar ratio profile as obtained with the non-amplified DNA. (c) X-chromosome evaluation of the 5-cell pool, again with a similar ratio profile. (d) X-chromosome evaluation of the single cell ‘#1’ from proband P1. For this cell the deletion on chromosome 22 was also identified.
Similar articles
- The use of ultra-dense array CGH analysis for the discovery of micro-copy number alterations and gene fusions in the cancer genome.
Przybytkowski E, Ferrario C, Basik M. Przybytkowski E, et al. BMC Med Genomics. 2011 Jan 27;4:16. doi: 10.1186/1755-8794-4-16. BMC Med Genomics. 2011. PMID: 21272361 Free PMC article. - Comprehensive performance comparison of high-resolution array platforms for genome-wide Copy Number Variation (CNV) analysis in humans.
Haraksingh RR, Abyzov A, Urban AE. Haraksingh RR, et al. BMC Genomics. 2017 Apr 24;18(1):321. doi: 10.1186/s12864-017-3658-x. BMC Genomics. 2017. PMID: 28438122 Free PMC article. - Technical demonstration of whole genome array comparative genomic hybridization.
Kennett JY, Watson SK, Saprunoff H, Heryet C, Lam WL. Kennett JY, et al. J Vis Exp. 2008 Aug 5;(18):870. doi: 10.3791/870. J Vis Exp. 2008. PMID: 19066503 Free PMC article. - Statistical issues in the analysis of DNA Copy Number Variations.
Wineinger NE, Kennedy RE, Erickson SW, Wojczynski MK, Bruder CE, Tiwari HK. Wineinger NE, et al. Int J Comput Biol Drug Des. 2008;1(4):368-95. doi: 10.1504/IJCBDD.2008.022208. Int J Comput Biol Drug Des. 2008. PMID: 19774103 Free PMC article. Review. - Novel applications of array comparative genomic hybridization in molecular diagnostics.
Cheung SW, Bi W. Cheung SW, et al. Expert Rev Mol Diagn. 2018 Jun;18(6):531-542. doi: 10.1080/14737159.2018.1479253. Epub 2018 May 31. Expert Rev Mol Diagn. 2018. PMID: 29848116 Review.
Cited by
- Non-invasive detection of genome-wide somatic copy number alterations by liquid biopsies.
Heitzer E, Ulz P, Geigl JB, Speicher MR. Heitzer E, et al. Mol Oncol. 2016 Mar;10(3):494-502. doi: 10.1016/j.molonc.2015.12.004. Epub 2015 Dec 17. Mol Oncol. 2016. PMID: 26778171 Free PMC article. Review. - New array approaches to explore single cells genomes.
Vanneste E, Bittman L, Van der Aa N, Voet T, Vermeesch JR. Vanneste E, et al. Front Genet. 2012 Mar 27;3:44. doi: 10.3389/fgene.2012.00044. eCollection 2012. Front Genet. 2012. PMID: 22509179 Free PMC article. - Genetic and epigenetic analysis of putative breast cancer stem cell models.
Balic M, Schwarzenbacher D, Stanzer S, Heitzer E, Auer M, Geigl JB, Cote RJ, Datar RH, Dandachi N. Balic M, et al. BMC Cancer. 2013 Jul 24;13:358. doi: 10.1186/1471-2407-13-358. BMC Cancer. 2013. PMID: 23883436 Free PMC article. - Single cell genomics: advances and future perspectives.
Macaulay IC, Voet T. Macaulay IC, et al. PLoS Genet. 2014 Jan 30;10(1):e1004126. doi: 10.1371/journal.pgen.1004126. eCollection 2014 Jan. PLoS Genet. 2014. PMID: 24497842 Free PMC article. Review. - Modeling genome coverage in single-cell sequencing.
Daley T, Smith AD. Daley T, et al. Bioinformatics. 2014 Nov 15;30(22):3159-65. doi: 10.1093/bioinformatics/btu540. Epub 2014 Aug 8. Bioinformatics. 2014. PMID: 25107873 Free PMC article.
References
- Iafrate AJ, Feuk L, Rivera MN, Listewnik ML, Donahoe PK, Qi Y, Scherer SW, Lee C. Detection of large-scale variation in the human genome. Nat. Genet. 2004;36:949–951. - PubMed
- Sebat J, Lakshmi B, Troge J, Alexander J, Young J, Lundin P, Maner S, Massa H, Walker M, Chi M, et al. Large-scale copy number polymorphism in the human genome. Science. 2004;305:525–528. - PubMed
- Tuzun E, Sharp AJ, Bailey JA, Kaul R, Morrison VA, Pertz LM, Haugen E, Hayden H, Albertson D, Pinkel D, et al. Fine-scale structural variation of the human genome. Nat. Genet. 2005;37:727–732. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous