Epigenetic profiling at mouse imprinted gene clusters reveals novel epigenetic and genetic features at differentially methylated regions - PubMed (original) (raw)
Epigenetic profiling at mouse imprinted gene clusters reveals novel epigenetic and genetic features at differentially methylated regions
Scott V Dindot et al. Genome Res. 2009 Aug.
Abstract
Genomic imprinting arises from allele-specific epigenetic modifications that are established during gametogenesis and that are maintained throughout somatic development. These parental-specific modifications include DNA methylation and post-translational modifications to histones, which create allele-specific active and repressive domains at imprinted regions. Through the use of a high-density genomic tiling array, we generated DNA and histone methylation profiles at 11 imprinted gene clusters in the mouse from DNA and from chromatin immunoprecipitated from sperm, heart, and cerebellum. Our analysis revealed that despite high levels of differential DNA methylation at non-CpG islands within these regions, imprinting control regions (ICRs) and secondary differentially methylated regions (DMRs) were identified by an overlapping pattern of H3K4 trimethylation (active chromatin) and H3K9 trimethylation (repressive chromatin) modifications in somatic tissue, and a sperm differentially methylated region (sDMR; sperm not equal somatic tissue). Using these features as a common signature of DMRs, we identified 11 unique regions that mapped to known imprinted genes, to uncharacterized genes, and to intergenic regions flanking known imprinted genes. A common feature among these regions was the presence of a CpG island and an array of tandem repeats. Collectively, this study provides a comprehensive analysis of DNA methylation and histone H3K4me3 and H3K9me3 modifications at imprinted gene clusters, and identifies common epigenetic and genetic features of regions regulating genomic imprinting.
Figures
Figure 1.
DNA methylation patterns at imprinted gene clusters. (A) MeDIP was performed on DNA isolated from sperm, heart, and cerebellum and the enriched (positive log2 IP/IN) values are plotted for each imprinted gene cluster. Chromosomal regions are marked as black boxes with corresponding chromosome numbers. (B) Sequence characterization of methylated regions in sperm, heart, and cerebellum showed that the imprinted gene clusters were primarily methylated at repetitive DNA based on repeat masker. (C) Plots showing the genomic location of methylated regions relative to the RefSeq gene annotation. (D) CpG island methylation was less frequent in sperm samples compared to the heart and cerebellum, particularly for the promoter and intragenic regions. (E) The majority of methylated CpG islands in heart and cerebellum were present in the promoters of known imprinted genes.
Figure 2.
Characterization of sperm-specific and tissue-specific differentially methylated regions at imprinted gene clusters. (A) A sliding window _t_-test was performed between sperm and heart (Sp vs. Ht; sDMR), sperm and cerebellum (Sp vs. Cb; sDMR), and heart and cerebellum (Ht vs. Cb; tDMR) using the log2 IP/IN ratios obtained from MeDIP to determine sperm and tissue-specific differentially methylated regions (sDMR and tDMR). The _y_-axis indicates regions that have significantly different (P < 0.00001) methylation profiles between samples and the direction of methylation. (B) The mean log2 IP/IN of probes was calculated over each known DMR in cerebellum, heart, and sperm. Box plots show less methylation in sperm DNA at each DMR, except at the H19 ICR and IG-DMR, which are methylated in sperm and differentially methylated in heart and cerebellum. Box plots represent the 25th and 75th percentile for each averaged sample. Whiskers show 10th and 90th percentiles. (C) An example showing the different patterns observed between sperm DNA methylation (Sperm DNAme) and heart DNA methylation (Heart DNAme) at the Mest DMR. (D) Sequence characterization of sDMR and tDMR showed that nonrepetitive DNA is primarily differentially methylated at imprinted gene clusters. (E) Genomic location of sDMRs and tDMRs relative to the RefSeq gene annotation. (F) An example of a tDMR at the 3′ end of the Igf2 gene. (G) Bisulfite sequencing confirmed loss of methylation in the cerebellum DNA at the 3′ end of the Igf2 gene. Arrows in F indicate regions amplified for bisulfite sequencing.
Figure 3.
H3K4me3 and H3K9me3 enriched regions at imprinted gene clusters. (A) ChIP-chip profiles of histone modifications for H3K4me3 and H3K9me3 in heart and cerebellum. _Y_-axis values represent log2 IP/IN. (B) Sequence characterization of regions enriched for H3K4me3 and H3K9me3 showed that each modification was present primarily at repetitive sequences. (C) Genomic location of H3K4me3 and H3K4me3 modifications relative to the RefSeq gene annotation. (D) Venn diagram showing overlapping H3K4me3 and H3K9me3 regions in both heart and cerebellum. (E) Genomic locations of overlapping H3K4me3 and H3K9me3 modifications relative to the RefSeq gene annotations.
Figure 4.
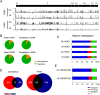
Overlapping patterns of H3K4me3 and H3K9me3 at sDMRs identified imprinting control regions. (A) An example of the Igf2r DMR in intron 2 of the Igf2r locus. The CpG island (green box) indicates the DMR, which contains both H3K4me3 and H3K9me3 modifications and differentially methylated DNA between sperm and heart. (B) Examples of the Ndn and Magel2 DMR showing overlapping H3K4me3 and H3K9me3, as well as sDMR between sperm and heart. (C) Genomic locations of regions with a sDMR and overlapping H3K4me3 and H3K9me3. (D) Genomic locations of CpG islands with a sDMR and overlapping H3K4me3 and H3K9me3. (E) Sequence characterization of regions with a sDMR and overlapping H3K4me3 and H3K9me3 indicated regions are highly repetitive and (F) are comprised mostly of simple repeats, SINES, and low complexity repeats. (G) Examples of sequence alignments for known ICRs (e.g., Igf2r DMR, Meg3 DMR, H19 ICR, and Tsix) and putative ICRs (e.g., Olfr1349, Upstream Ndn, 5730403M16Rik, and Xlr5C) showing repeated elements.
Similar articles
- Coordinated allele-specific histone acetylation at the differentially methylated regions of imprinted genes.
Singh P, Cho J, Tsai SY, Rivas GE, Larson GP, Szabó PE. Singh P, et al. Nucleic Acids Res. 2010 Dec;38(22):7974-90. doi: 10.1093/nar/gkq680. Epub 2010 Aug 6. Nucleic Acids Res. 2010. PMID: 20693536 Free PMC article. - A genome-wide screen for normally methylated human CpG islands that can identify novel imprinted genes.
Strichman-Almashanu LZ, Lee RS, Onyango PO, Perlman E, Flam F, Frieman MB, Feinberg AP. Strichman-Almashanu LZ, et al. Genome Res. 2002 Apr;12(4):543-54. doi: 10.1101/gr.224102. Genome Res. 2002. PMID: 11932239 Free PMC article. - Hemimethylation of CpG dyads is characteristic of secondary DMRs associated with imprinted loci and correlates with 5-hydroxymethylcytosine at paternally methylated sequences.
Nechin J, Tunstall E, Raymond N, Hamagami N, Pathmanabhan C, Forestier S, Davis TL. Nechin J, et al. Epigenetics Chromatin. 2019 Oct 17;12(1):64. doi: 10.1186/s13072-019-0309-2. Epigenetics Chromatin. 2019. PMID: 31623686 Free PMC article. - Imprinting of the mouse Igf2r gene depends on an intronic CpG island.
Wutz A, Barlow DP. Wutz A, et al. Mol Cell Endocrinol. 1998 May 25;140(1-2):9-14. doi: 10.1016/s0303-7207(98)00022-7. Mol Cell Endocrinol. 1998. PMID: 9722161 Review. - How genome-wide approaches can be used to unravel the remaining secrets of the imprintome.
Cooper WN, Constância M. Cooper WN, et al. Brief Funct Genomics. 2010 Jul;9(4):315-28. doi: 10.1093/bfgp/elq018. Brief Funct Genomics. 2010. PMID: 20675687 Review.
Cited by
- Astrocytic ALKBH5 in stress response contributes to depressive-like behaviors in mice.
Guo F, Fan J, Liu JM, Kong PL, Ren J, Mo JW, Lu CL, Zhong QL, Chen LY, Jiang HT, Zhang C, Wen YL, Gu TT, Li SJ, Fang YY, Pan BX, Gao TM, Cao X. Guo F, et al. Nat Commun. 2024 May 21;15(1):4347. doi: 10.1038/s41467-024-48730-2. Nat Commun. 2024. PMID: 38773146 Free PMC article. - Coordinated changes in AHRR methylation in lymphoblasts and pulmonary macrophages from smokers.
Monick MM, Beach SR, Plume J, Sears R, Gerrard M, Brody GH, Philibert RA. Monick MM, et al. Am J Med Genet B Neuropsychiatr Genet. 2012 Mar;159B(2):141-51. doi: 10.1002/ajmg.b.32021. Epub 2012 Jan 9. Am J Med Genet B Neuropsychiatr Genet. 2012. PMID: 22232023 Free PMC article. - A DNA methylation fingerprint of 1628 human samples.
Fernandez AF, Assenov Y, Martin-Subero JI, Balint B, Siebert R, Taniguchi H, Yamamoto H, Hidalgo M, Tan AC, Galm O, Ferrer I, Sanchez-Cespedes M, Villanueva A, Carmona J, Sanchez-Mut JV, Berdasco M, Moreno V, Capella G, Monk D, Ballestar E, Ropero S, Martinez R, Sanchez-Carbayo M, Prosper F, Agirre X, Fraga MF, Graña O, Perez-Jurado L, Mora J, Puig S, Prat J, Badimon L, Puca AA, Meltzer SJ, Lengauer T, Bridgewater J, Bock C, Esteller M. Fernandez AF, et al. Genome Res. 2012 Feb;22(2):407-19. doi: 10.1101/gr.119867.110. Epub 2011 May 25. Genome Res. 2012. PMID: 21613409 Free PMC article. - PHF13 is a molecular reader and transcriptional co-regulator of H3K4me2/3.
Chung HR, Xu C, Fuchs A, Mund A, Lange M, Staege H, Schubert T, Bian C, Dunkel I, Eberharter A, Regnard C, Klinker H, Meierhofer D, Cozzuto L, Winterpacht A, Di Croce L, Min J, Will H, Kinkley S. Chung HR, et al. Elife. 2016 May 25;5:e10607. doi: 10.7554/eLife.10607. Elife. 2016. PMID: 27223324 Free PMC article. - Targeted DNA methylation analysis by high throughput sequencing in porcine peri-attachment embryos.
Morrill BH, Cox L, Ward A, Heywood S, Prather RS, Isom SC. Morrill BH, et al. J Reprod Dev. 2013;59(3):314-20. doi: 10.1262/jrd.2012-144. Epub 2013 Feb 22. J Reprod Dev. 2013. PMID: 23428632 Free PMC article.
References
- Arima T, Yamasaki K, John RM, Kato K, Sakumi K, Nakabeppu Y, Wake N, Kono T. The human HYMAI/PLAGL1 differentially methylated region acts as an imprint control region in mice. Genomics. 2006;88:650–658. - PubMed
- Delaval K, Feil R. Epigenetic regulation of mammalian genomic imprinting. Curr Opin Genet Dev. 2004;14:188–195. - PubMed
- Edwards CA, Ferguson-Smith AC. Mechanisms regulating imprinted genes in clusters. Curr Opin Cell Biol. 2007;19:281–289. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases