PCSIM: A Parallel Simulation Environment for Neural Circuits Fully Integrated with Python - PubMed (original) (raw)
PCSIM: A Parallel Simulation Environment for Neural Circuits Fully Integrated with Python
Dejan Pecevski et al. Front Neuroinform. 2009.
Abstract
The Parallel Circuit SIMulator (PCSIM) is a software package for simulation of neural circuits. It is primarily designed for distributed simulation of large scale networks of spiking point neurons. Although its computational core is written in C++, PCSIM's primary interface is implemented in the Python programming language, which is a powerful programming environment and allows the user to easily integrate the neural circuit simulator with data analysis and visualization tools to manage the full neural modeling life cycle. The main focus of this paper is to describe PCSIM's full integration into Python and the benefits thereof. In particular we will investigate how the automatically generated bidirectional interface and PCSIM's object-oriented modular framework enable the user to adopt a hybrid modeling approach: using and extending PCSIM's functionality either employing pure Python or C++ and thus combining the advantages of both worlds. Furthermore, we describe several supplementary PCSIM packages written in pure Python and tailored towards setting up and analyzing neural simulations.
Keywords: Boost.Python; PCSIM; Py++; Python; neural simulator; parallel simulation; spiking neurons.
Figures
Figure 1
Architecture overview of PCSIM.
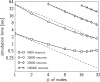
Figure 2
Simulation times of the CUBA network distributed over different number of processing nodes, compared to the expected simulation time (dashed line) (see text for details). Four different sizes of networks were simulated: 4000 neurons with on average 1.6 × 106 synapses (squares), 20000 neurons with on verage 40 × 106 synapses (circles), 50000 neurons with on average 250 × 106 synapses (diamonds) and 100000 neurons with on average 1 × 109 synapses (crosses). The plotted simulation times are averages over 12 simulation runs. The variation of simulation time between different simulation runs was small, therefore we did not show it.
Figure 3
The processing steps in the generation of the Python interface for PCSIM.
Figure 4
A diagram of the most important concepts within the network construction interface. The arrows indicate a “uses” relationship between the concepts.
Figure 5
(A) Network elements of different type (with different arrangement of input and output ports) interconnected together in a PCSIM network. Different colors of ports, gray or white, mark their different types, spiking or analog. (B) Neurons and synapses are specific subtypes of the more general concept of an network element. (C) Schematic diagram of the embedding of a network simulated with the Brian simulator into a PCSIM network element.
Figure 6
Plots from the output analysis example with the pypcsimplus package. (A) Spike response of the spiking network implemented in the Section “Custom Network Elements”, with input neurons emitting spikes generated from a homogeneous Poisson process with a rate of 5 Hz, for the first 0.4 s of the simulation. (B) Cross-correlogram of the spike response of the network model from (A). (C) Spike response of the spiking network implemented in the Section “Custom Network Elements”, when the input neurons emit spikes generated from an inhomogeneous Poisson process with a rate changing according to a sinusoidal function (see text for details). (D) Cross-correlogram of the spike response of the network model from (C).
Similar articles
- NEVESIM: event-driven neural simulation framework with a Python interface.
Pecevski D, Kappel D, Jonke Z. Pecevski D, et al. Front Neuroinform. 2014 Aug 14;8:70. doi: 10.3389/fninf.2014.00070. eCollection 2014. Front Neuroinform. 2014. PMID: 25177291 Free PMC article. - Brainlab: A Python Toolkit to Aid in the Design, Simulation, and Analysis of Spiking Neural Networks with the NeoCortical Simulator.
Drewes R, Zou Q, Goodman PH. Drewes R, et al. Front Neuroinform. 2009 May 27;3:16. doi: 10.3389/neuro.11.016.2009. eCollection 2009. Front Neuroinform. 2009. PMID: 19506707 Free PMC article. - PyNEST: A Convenient Interface to the NEST Simulator.
Eppler JM, Helias M, Muller E, Diesmann M, Gewaltig MO. Eppler JM, et al. Front Neuroinform. 2009 Jan 29;2:12. doi: 10.3389/neuro.11.012.2008. eCollection 2008. Front Neuroinform. 2009. PMID: 19198667 Free PMC article. - GPUPeP: Parallel Enzymatic Numerical P System simulator with a Python-based interface.
Raghavan S, Rai SS, Rohit MP, Chandrasekaran K. Raghavan S, et al. Biosystems. 2020 Oct;196:104186. doi: 10.1016/j.biosystems.2020.104186. Epub 2020 Jun 11. Biosystems. 2020. PMID: 32535178 Review. - Simulation of networks of spiking neurons: a review of tools and strategies.
Brette R, Rudolph M, Carnevale T, Hines M, Beeman D, Bower JM, Diesmann M, Morrison A, Goodman PH, Harris FC Jr, Zirpe M, Natschläger T, Pecevski D, Ermentrout B, Djurfeldt M, Lansner A, Rochel O, Vieville T, Muller E, Davison AP, El Boustani S, Destexhe A. Brette R, et al. J Comput Neurosci. 2007 Dec;23(3):349-98. doi: 10.1007/s10827-007-0038-6. Epub 2007 Jul 12. J Comput Neurosci. 2007. PMID: 17629781 Free PMC article. Review.
Cited by
- NeuroML: a language for describing data driven models of neurons and networks with a high degree of biological detail.
Gleeson P, Crook S, Cannon RC, Hines ML, Billings GO, Farinella M, Morse TM, Davison AP, Ray S, Bhalla US, Barnes SR, Dimitrova YD, Silver RA. Gleeson P, et al. PLoS Comput Biol. 2010 Jun 17;6(6):e1000815. doi: 10.1371/journal.pcbi.1000815. PLoS Comput Biol. 2010. PMID: 20585541 Free PMC article. - Pyff - a pythonic framework for feedback applications and stimulus presentation in neuroscience.
Venthur B, Scholler S, Williamson J, Dähne S, Treder MS, Kramarek MT, Müller KR, Blankertz B. Venthur B, et al. Front Neurosci. 2010 Dec 2;4:179. doi: 10.3389/fnins.2010.00179. eCollection 2010. Front Neurosci. 2010. PMID: 21160550 Free PMC article. - Meeting the memory challenges of brain-scale network simulation.
Kunkel S, Potjans TC, Eppler JM, Plesser HE, Morrison A, Diesmann M. Kunkel S, et al. Front Neuroinform. 2012 Jan 24;5:35. doi: 10.3389/fninf.2011.00035. eCollection 2011. Front Neuroinform. 2012. PMID: 22291636 Free PMC article. - NFTsim: Theory and Simulation of Multiscale Neural Field Dynamics.
Sanz-Leon P, Robinson PA, Knock SA, Drysdale PM, Abeysuriya RG, Fung FK, Rennie CJ, Zhao X. Sanz-Leon P, et al. PLoS Comput Biol. 2018 Aug 22;14(8):e1006387. doi: 10.1371/journal.pcbi.1006387. eCollection 2018 Aug. PLoS Comput Biol. 2018. PMID: 30133448 Free PMC article. - Stochastic computations in cortical microcircuit models.
Habenschuss S, Jonke Z, Maass W. Habenschuss S, et al. PLoS Comput Biol. 2013;9(11):e1003311. doi: 10.1371/journal.pcbi.1003311. Epub 2013 Nov 14. PLoS Comput Biol. 2013. PMID: 24244126 Free PMC article.
References
- Abrahams D., Grosse-Kunstleve R. W. (2003). Building hybrid systems with Boost.Python. C/C++ Users J. 21, 29–36
- Beazley D. (2003). Automated scientific software scripting with SWIG. Future Generat. Comput. Syst. 19, 599–60910.1016/S0167-739X(02)00171-1 - DOI
- Bower J. M., Beeman D. (1998). The Book of GENESIS (2nd ed.): Exploring Realistic Neural Models With the GEneral NEural SImulation System. New York, Springer-Verlag New York, Inc.
- Brette R., Rudolph M., Carnevale T., Hines M., Beeman D., Bower J. M., Diesmann M., Morrison A., Goodman P. H., Harris F. C., Jr, Zirpe M., Natschläger T., Pecevski D., Ermentrout B., Djurfeldt M., Lansner A., Rochel O., Vieville T., Muller E., Davison A. P., Boustani S. E., Destexhe A. (2007). Simulation of networks of spiking neurons: a review of tools and strategies. J. Comput. Neurosci. 23, 349–39810.1007/s10827-007-0038-6 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources