Oncosome formation in prostate cancer: association with a region of frequent chromosomal deletion in metastatic disease - PubMed (original) (raw)
. 2009 Jul 1;69(13):5601-9.
doi: 10.1158/0008-5472.CAN-08-3860. Epub 2009 Jun 23.
Jayoung Kim, Martin H Hager, Matteo Morello, Wei Yang, Christopher J Lafargue, Lawrence D True, Mark A Rubin, Rosalyn M Adam, Rameen Beroukhim, Francesca Demichelis, Michael R Freeman
Affiliations
- PMID: 19549916
- PMCID: PMC2853876
- DOI: 10.1158/0008-5472.CAN-08-3860
Oncosome formation in prostate cancer: association with a region of frequent chromosomal deletion in metastatic disease
Dolores Di Vizio et al. Cancer Res. 2009.
Abstract
Oncosomes have recently been described as membrane-derived microvesicles secreted by cancer cells, which transfer oncogenic signals and protein complexes across cell boundaries. Here, we show the rapid formation and secretion of oncosomes from DU145 and LNCaP human prostate cancer cells. Oncosome formation was stimulated by epidermal growth factor receptor activation and also by overexpression of membrane-targeted Akt1. Microvesicles shed from prostate cancer cells contained numerous signal transduction proteins and were capable of activating rapid phospho-tyrosine and Akt pathway signaling, and stimulating proliferation and migration, in recipient tumor cells. They also induced a stromal reaction in recipient normal cells. Knockdown of the actin nucleating protein Diaphanous Related Formin 3 (DRF3/Dia2) by RNA interference enhanced rates of oncosome formation, indicating that these structures resemble, and may be identical to, nonapoptotic membrane blebs, a feature of the amoeboid form of cell motility. Analysis of primary and metastatic human prostate tumors using 100K single nucleotide polymorphism arrays revealed a significantly higher frequency of deletion of the locus encoding DRF3 (DIAPH3) in metastatic tumors (P = 0.001) in comparison with organ-confined tumors. Fluorescence in situ hybridization confirmed increased chromosomal loss of DIAPH3 in metastatic tumors in a different cohort of patients (P = 0.006). These data suggest that microvesicles shed from prostate cancer cells can alter the tumor microenvironment in a manner that may promote disease progression. They also show that DRF3 is a physiologically relevant protein that seems to regulate this process.
Figures
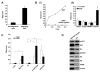
Figure 1. EGF induces formation of non-apoptotic membrane blebs
(A) Membrane staining with 0.5 μg/ml FITC-CTxB for 5 min, after 3 h treatment with: (−) vehicle or (+) EGF (50 ng/ml), and imaged by confocal microscopy (63X). All panels show DU145 cells, except the panel at upper right, which shows a PC-3 cell and vesicles shed (SV) into the medium. The inset shows a single, attached bleb at higher power. (B) Frames (5 sec interval) from Movies 1 and 2 acquired by real time confocal microscopy of DU145 cells expressing membrane-targeted pMEM-YFP, and treated with EGF. Panel B shows two examples (differently shaped arrows) of bleb dynamics. (C) Lysates from whole cells and SV contain endogenous Cav-1, consistent with localization of a Cav-1-GFP fusion to membrane blebs. (D) EGF does not induce apoptosis under these experimental conditions, as shown by an assay for cleaved PARP.
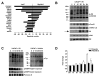
Figure 2. Blebbing is associated with cell migration, activation of signaling and results in production of shed vesicles that contain membrane proteins
(A) Bleb formation in DU145 cells treated with EGF (50 ng/ml, 12 h) in a wound healing assay. (B) (i) Bleb formation in DU145 cells treated with EGF or EGF plus gefitinib (ZD1839) (10 μM). (ii) Bleb formation in prostate normal epithelial (iPrEC) and stromal (WPMY-1) cells, in comparison with DU145, with and without EGF. iPrECa were kept in PrEGM medium, which is serum-free but contains EGF, while iPrECb were transferred to serum-free RPMI 12 h before treatment. (C) Bleb formation in LNCaP cells stably engineered to express MyrAkt1, pro-HB-EGF, or soluble (constitutively secreted) sHB-EGF. LacZ (irrelevant gene) and Vo (vector only) are negative controls. (D) Protein from whole cell lysates and SV from EGF-treated LNCaP/MyrAkt1 cells were blotted with the indicated Abs.
Figure 3. Shed vesicles exhibit oncosome activity
(A) Proteins identified by tandem mass spectrometry in bleb material and quantified using spectral counting. PDCD6IP (Programmed Cell Death 6 Interacting Protein), PABPC1 (Poly(A) Binding Protein, Cytoplasmic 1), HSPA8 (Heat Shock 70kDa Protein 8), ANXA6 (Annexin A 6), hnRNP-K (Heterogeneous Nuclear Ribonucleoprotein K, PKM2 (Pyruvate Kinase M2), PACSIN2 (Protein Kinase C and Casein Kinase Substrate in Neurons 2), PABPC4 (Poly(A) Binding Protein, Cytoplasmic 4), RPLPO (Ribosomal Protein Large P0-like Protein), IGSF8 (Immunoglobulin Superfamily, member 8), YWHAE (Tyrosine 3-monooxygenase/tryptophan 5-monooxygenase Activation Protein, Epsilon Polypeptide), HSPA1L (Heat Shock 70kDa Protein 1-Like), RPL7 (Ribosomal Protein L7), HSPA1A (Heat Shock 70kDa protein 1A), S100A7 (S100 calcium binding protein A7), JUP (Junction Plakoglobin), XRCC6 (X-ray Repair Complementing defective repair in Chinese hamster cells 6). (B) LNCaP/LacZ cells were exposed to SV obtained from EGF-treated LNCaP/MyrAkt1 cells. Blotting of whole cell lysates is shown. (C) LNCaP/LacZ exposed to 20 μg of SV or vehicle from EGF-treated LNCaP/MyrAkt1 cells and assessed for p-Tyr (upper panels). The lower panels show the results of blotting with p-EGFR antibody (p-Tyr1068). (D) WPMY-1 cells were exposed to LNCaP/MyrAkt1 derived SV for the indicated times. Vimentin mRNA was quantified by qRT-PCR in recipient cells, and normalized using GAPDH. An irrelevant gene, PRKAR1A, was used as control.
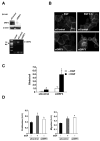
Figure 4. DRF3 knockdown by RNA interference results in oncosome secretion
(A) Verification of DIAPH3 gene silencing by siDRF3 in DU145 cells by western blot (upper panel) and RT-PCR (lower panel). Control non-targeting siRNA was a negative control. (B) FITC-CTxB staining of DU145 cells showing blebbing in siDRF3-transfected or siRNA control cells, with or without EGF (3 h) (right panel). (C) Quantitative analysis of bleb formation in DU145 cells treated with siRNA for DRF3 or control siRNA, +/− EGF. (D) Proliferation assay (left panel) and migration assay (right panel) in DU145 cells treated with SV prepared from DU145 cells treated with siRNA control oligos, +/− EGF, or with DRF3-targeted oligos. *p<0.05.
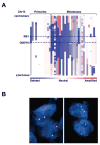
Figure 5. Genomic profiling of primary and metastatic prostate cancer at the DIAPH3 locus
(A) Amplifications (red) and deletions (blue), determined by segmentation analysis of normalized signal intensities from 100K SNP arrays (see Methods), are displayed for 39 prostate cancers (x-axis; primaries and metastases are designated along the top) for the q arm of chromosome 13 (chromosomal positions indicated along the y-axis include the centromere, q-terminus, and RB1 loci). (B) DIAPH3 fluorescent in situ hybridization was performed by dual color FISH on PCa tissues. The FISH image on the left shows both red signals (DIAPH3 locus on chr13q21.2) and green signals (a stable region on chr21q22.12) in representative nuclei indicating no deletion of DIAPH3 in tumor cells. The FISH image on the right shows one red signal (DIAPH3 locus on chr13q21.2) and two green signals (a stable region on chr21q22.12) in representative nuclei. Loss of the second red signal is consistent with deletion at DIAPH3. Original magnification of FISH images, 60 X objective.
Similar articles
- Enhanced shedding of extracellular vesicles from amoeboid prostate cancer cells: potential effects on the tumor microenvironment.
Kim J, Morley S, Le M, Bedoret D, Umetsu DT, Di Vizio D, Freeman MR. Kim J, et al. Cancer Biol Ther. 2014 Apr;15(4):409-18. doi: 10.4161/cbt.27627. Epub 2014 Jan 14. Cancer Biol Ther. 2014. PMID: 24423651 Free PMC article. - ASAP1, a gene at 8q24, is associated with prostate cancer metastasis.
Lin D, Watahiki A, Bayani J, Zhang F, Liu L, Ling V, Sadar MD, English J, Fazli L, So A, Gout PW, Gleave M, Squire JA, Wang YZ. Lin D, et al. Cancer Res. 2008 Jun 1;68(11):4352-9. doi: 10.1158/0008-5472.CAN-07-5237. Cancer Res. 2008. PMID: 18519696 - RNA interference-directed knockdown of urokinase plasminogen activator and urokinase plasminogen activator receptor inhibits prostate cancer cell invasion, survival, and tumorigenicity in vivo.
Pulukuri SM, Gondi CS, Lakka SS, Jutla A, Estes N, Gujrati M, Rao JS. Pulukuri SM, et al. J Biol Chem. 2005 Oct 28;280(43):36529-40. doi: 10.1074/jbc.M503111200. Epub 2005 Aug 26. J Biol Chem. 2005. PMID: 16127174 Free PMC article. Retracted. - DIAPH3 governs the cellular transition to the amoeboid tumour phenotype.
Hager MH, Morley S, Bielenberg DR, Gao S, Morello M, Holcomb IN, Liu W, Mouneimne G, Demichelis F, Kim J, Solomon KR, Adam RM, Isaacs WB, Higgs HN, Vessella RL, Di Vizio D, Freeman MR. Hager MH, et al. EMBO Mol Med. 2012 Aug;4(8):743-60. doi: 10.1002/emmm.201200242. Epub 2012 May 16. EMBO Mol Med. 2012. PMID: 22593025 Free PMC article. - Prostate cancer--biology of metastasis and its clinical implications.
Dong JT, Rinker-Schaeffer CW, Ichikawa T, Barrett JC, Isaacs JT. Dong JT, et al. World J Urol. 1996;14(3):182-9. doi: 10.1007/BF00186898. World J Urol. 1996. PMID: 8806197 Review.
Cited by
- Decoding the secret of extracellular vesicles in the immune tumor microenvironment of the glioblastoma: on the border of kingdoms.
Ghazi B, Harmak Z, Rghioui M, Kone AS, El Ghanmi A, Badou A. Ghazi B, et al. Front Immunol. 2024 Aug 29;15:1423232. doi: 10.3389/fimmu.2024.1423232. eCollection 2024. Front Immunol. 2024. PMID: 39267734 Free PMC article. Review. - Regulation of microtubule dynamics by DIAPH3 influences amoeboid tumor cell mechanics and sensitivity to taxanes.
Morley S, You S, Pollan S, Choi J, Zhou B, Hager MH, Steadman K, Spinelli C, Rajendran K, Gertych A, Kim J, Adam RM, Yang W, Krishnan R, Knudsen BS, Di Vizio D, Freeman MR. Morley S, et al. Sci Rep. 2015 Jul 16;5:12136. doi: 10.1038/srep12136. Sci Rep. 2015. PMID: 26179371 Free PMC article. - Uptake and Fate of Extracellular Membrane Vesicles: Nucleoplasmic Reticulum-Associated Late Endosomes as a New Gate to Intercellular Communication.
Corbeil D, Santos MF, Karbanová J, Kurth T, Rappa G, Lorico A. Corbeil D, et al. Cells. 2020 Aug 21;9(9):1931. doi: 10.3390/cells9091931. Cells. 2020. PMID: 32825578 Free PMC article. Review. - Large oncosomes overexpressing integrin alpha-V promote prostate cancer adhesion and invasion via AKT activation.
Ciardiello C, Leone A, Lanuti P, Roca MS, Moccia T, Minciacchi VR, Minopoli M, Gigantino V, De Cecio R, Rippa M, Petti L, Capone F, Vitagliano C, Milone MR, Pucci B, Lombardi R, Iannelli F, Di Gennaro E, Bruzzese F, Marchisio M, Carriero MV, Di Vizio D, Budillon A. Ciardiello C, et al. J Exp Clin Cancer Res. 2019 Jul 18;38(1):317. doi: 10.1186/s13046-019-1317-6. J Exp Clin Cancer Res. 2019. PMID: 31319863 Free PMC article. - Large and small extracellular vesicles released by glioma cells in vitro and in vivo.
Yekula A, Minciacchi VR, Morello M, Shao H, Park Y, Zhang X, Muralidharan K, Freeman MR, Weissleder R, Lee H, Carter B, Breakefield XO, Di Vizio D, Balaj L. Yekula A, et al. J Extracell Vesicles. 2019 Nov 27;9(1):1689784. doi: 10.1080/20013078.2019.1689784. eCollection 2020. J Extracell Vesicles. 2019. PMID: 31839905 Free PMC article.
References
- Cao J, Chiarelli C, Richman O, Zarrabi K, Kozarekar P, Zucker S. Membrane type 1 matrix metalloproteinase induces epithelial-to-mesenchymal transition in prostate cancer. J Biol Chem. 2008;283:6232–40. - PubMed
- Wang HR, Zhang Y, Ozdamar B, et al. Regulation of cell polarity and protrusion formation by targeting RhoA for degradation. Science. 2003;302:1775–9. - PubMed
- Katoh H, Hiramoto K, Negishi M. Activation of Rac1 by RhoG regulates cell migration. J Cell Sci. 2006;119:56–65. - PubMed
- Paluch E, Sykes C, Prost J, Bornens M. Dynamic modes of the cortical actomyosin gel during cell locomotion and division. Trends Cell Biol. 2006;16:5–10. - PubMed
- Gadea G, Sanz-Moreno V, Self A, Godi A, Marshall CJ. DOCK10-mediated Cdc42 activation is necessary for amoeboid invasion of melanoma cells. Curr Biol. 2008;18:1456–65. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- K08CA122833/CA/NCI NIH HHS/United States
- CA97186/CA/NCI NIH HHS/United States
- R3747556/PHS HHS/United States
- P50 CA097186/CA/NCI NIH HHS/United States
- P01 CA085859/CA/NCI NIH HHS/United States
- P01CA085859/CA/NCI NIH HHS/United States
- R01 CA112303/CA/NCI NIH HHS/United States
- K99 CA131472/CA/NCI NIH HHS/United States
- P50 DK065298/DK/NIDDK NIH HHS/United States
- R01CA116337-01A1/CA/NCI NIH HHS/United States
- P50 DK65298/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous