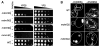An ER-mitochondria tethering complex revealed by a synthetic biology screen - PubMed (original) (raw)
An ER-mitochondria tethering complex revealed by a synthetic biology screen
Benoît Kornmann et al. Science. 2009.
Abstract
Communication between organelles is an important feature of all eukaryotic cells. To uncover components involved in mitochondria/endoplasmic reticulum (ER) junctions, we screened for mutants that could be complemented by a synthetic protein designed to artificially tether the two organelles. We identified the Mmm1/Mdm10/Mdm12/Mdm34 complex as a molecular tether between ER and mitochondria. The tethering complex was composed of proteins resident of both ER and mitochondria. With the use of genome-wide mapping of genetic interactions, we showed that the components of the tethering complex were functionally connected to phospholipid biosynthesis and calcium-signaling genes. In mutant cells, phospholipid biosynthesis was impaired. The tethering complex localized to discrete foci, suggesting that discrete sites of close apposition between ER and mitochondria facilitate interorganelle calcium and phospholipid exchange.
Figures
Fig. 1
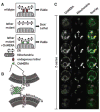
A synthetic biology screen to uncover mutants of the ER-mitochondria connection. (A) Rationale of the screen. (Top) In WT cells a yet unknown endogenous complex tethers the ER to the mitochondria. (Middle) Mutations causing the loss of the endogenous complex are detrimental and cause slow growth or cell death. (Bottom) Artificial ER-mitochondria tethering by ChiMERA can suppress the defects associated with the loss of the endogenous tether. (B) Outline of the ChiMERA. A central GFP molecule (green) is flanked by the mitochondria-directed N-terminal Tom70 presequence and transmembrane sequence (red) and the ER-directed C-terminal Ubc6 tail anchor sequence (blue). (C) Confocal Z-series across a yeast cell expressing the ChiMERA and a mitochondrial marker (mt-dsRed). ChiMERA displays a characteristic ER staining with additional thicker structures (arrowheads), which colocalize with mitochondria and represent sites of artificial tethering.
Fig. 2
ChiMERA expression suppresses phenotypes associated with deletions of ERMES complex components. (A) 10-fold serial dilutions of a saturated culture were spotted on YP + ethanol/glycerol medium (YPEG) that supports growth by respiration or YP + dextrose (YPD) that allows fermentation. ChiMERA expression rescues growth of mdm12Δ, mdm34Δ, mdm10Δ, and mmm1Δ strains to different extents on respiration medium. (B) The mitochondrial shape defect of mdm12Δ and mdm34Δ strains is partially suppressed by expression of the ChiMERA. Cells bearing a mitochondria-targeted dsRed were imaged with the use of confocal fluorescence microscopy. Z projections across the whole volume of the cells are shown. A broken white line outlines the perimeter of the cell. ChiMERA expression has no effect in WT cells, other than formation of artificial tethering structures. mdm12Δ and mdm34Δ strains display rounded and enlarged mitochondria. The mitochondrial tubular shape is substantially restored upon ChiMERA expression. A quantitation of this effect is shown in fig. S4.
Fig. 3
Mmm1 is an integral ER protein. (A) Disruption of a single ERMES component causes disassembly of the complex. Mmm1-GFP is localized in punctate structures in WT cells (a). Upon deletion of MDM12 (b), MDM34 (c), or MDM10 (d), Mmm1-GFP relocalizes to the ER. Mdm12-GFP displays a punctate pattern in WT cells (e). In the absence of Mmm1 (f), Mdm12-GFP displays a uniform mitochondrial localization (note the rounded swollen mitochondrial shape in mmm1Δ strains), but in the absence of MDM34 (g) or MDM10 (h), Mdm12-GFP relocalizes to the ER. Mdm34-GFP is also in punctate structures in WT cells (i). Upon mutation of any other complex member, Mdm34 relocalizes more uniformly to the mitochondrial membrane (j, k, l). Mdm10-GFP localizes to the whole surface of the mitochondria (m) and localizes to the rounded mitochondria after deletion of any other ERMES component (n, o, p). n.d., not determined. (B) Mmm1 is N-glycosylated. Whole-cell extract from a strain bearing a functional HA-tagged MMM1 gene was subjected to SDS–polyacrylamide gel electrophoresis with or without pretreatment with the glycosidase EndoHf. Detection was performed by Western blotting with an anti-HA antibody. The shift in electrophoretic mobility upon glycosidase treatment is indicative that Mmm1 is N-linked glycosylated. (C) Model of ERMES-mediated ER-mitochondria tethering. Mmm1 is an integral ER protein glycosylated on its N-terminal side. Mmm1 interacts with Mdm10, a OMM β-barrel protein. Mdm34 and Mdm12 promote this association, most probably via direct association.
Fig. 4
Global analysis of ERMES genetic interactions. (A) Genetic interactions of MMM1 and MDM10 (top) and of MDM12 and MDM34 (bottom). Positive values indicate epistatic or suppressive interactions (i.e., double mutant grows better than expected from the combination of the phenotypes of each single mutant); negative values indicate a synthetic sick/lethal genetic interaction (i.e., double mutant grows worse than expected). (B) Histograms of correlation coefficients generated by comparing the profiles of genetic interaction for each ERMES component to all other 1493 profiles in the E-MAP analysis. ERMES components display strongest correlation to each other and to GEM1 and PSD1. (C) Thin-layer chromatography (TLC) analysis of steady-state total and mitochondrial phospholipids in WT and mdm12Δ strains. PA, phosphatidic acid; PI, phosphatidylinositol. (D) The aminoglycerophospholipid biosynthesis pathway is slowed down in ERMES mutants. Cultures of the indicated genotypes were labeled with 14C-serine, then chased with an excess cold serine. The variation of the PC/PS ratio was then measured over time by quantitative TLC, and its slope was estimated with linear regression. Error bars represent the SE of the linear regression.
Comment in
- Cell biology. Connecting organelles.
Wiedemann N, Meisinger C, Pfanner N. Wiedemann N, et al. Science. 2009 Jul 24;325(5939):403-4. doi: 10.1126/science.1178016. Science. 2009. PMID: 19628848 No abstract available.
Similar articles
- Crystal structure of Mdm12 reveals the architecture and dynamic organization of the ERMES complex.
Jeong H, Park J, Lee C. Jeong H, et al. EMBO Rep. 2016 Dec;17(12):1857-1871. doi: 10.15252/embr.201642706. Epub 2016 Nov 7. EMBO Rep. 2016. PMID: 27821511 Free PMC article. - Structure-function insights into direct lipid transfer between membranes by Mmm1-Mdm12 of ERMES.
Kawano S, Tamura Y, Kojima R, Bala S, Asai E, Michel AH, Kornmann B, Riezman I, Riezman H, Sakae Y, Okamoto Y, Endo T. Kawano S, et al. J Cell Biol. 2018 Mar 5;217(3):959-974. doi: 10.1083/jcb.201704119. Epub 2017 Dec 26. J Cell Biol. 2018. PMID: 29279306 Free PMC article. - Crystal structures of Mmm1 and Mdm12-Mmm1 reveal mechanistic insight into phospholipid trafficking at ER-mitochondria contact sites.
Jeong H, Park J, Jun Y, Lee C. Jeong H, et al. Proc Natl Acad Sci U S A. 2017 Nov 7;114(45):E9502-E9511. doi: 10.1073/pnas.1715592114. Epub 2017 Oct 25. Proc Natl Acad Sci U S A. 2017. PMID: 29078410 Free PMC article. - The ERMES complex and ER-mitochondria connections.
Michel AH, Kornmann B. Michel AH, et al. Biochem Soc Trans. 2012 Apr;40(2):445-50. doi: 10.1042/BST20110758. Biochem Soc Trans. 2012. PMID: 22435828 Review. - Endoplasmic Reticulum-Mitochondria Communication Through Ca2+ Signaling: The Importance of Mitochondria-Associated Membranes (MAMs).
Marchi S, Bittremieux M, Missiroli S, Morganti C, Patergnani S, Sbano L, Rimessi A, Kerkhofs M, Parys JB, Bultynck G, Giorgi C, Pinton P. Marchi S, et al. Adv Exp Med Biol. 2017;997:49-67. doi: 10.1007/978-981-10-4567-7_4. Adv Exp Med Biol. 2017. PMID: 28815521 Review.
Cited by
- Tam41 is a CDP-diacylglycerol synthase required for cardiolipin biosynthesis in mitochondria.
Tamura Y, Harada Y, Nishikawa S, Yamano K, Kamiya M, Shiota T, Kuroda T, Kuge O, Sesaki H, Imai K, Tomii K, Endo T. Tamura Y, et al. Cell Metab. 2013 May 7;17(5):709-18. doi: 10.1016/j.cmet.2013.03.018. Epub 2013 Apr 25. Cell Metab. 2013. PMID: 23623749 Free PMC article. - Proteins that carry dual targeting signals can act as tethers between peroxisomes and partner organelles.
Bittner E, Stehlik T, Lam J, Dimitrov L, Heimerl T, Schöck I, Harberding J, Dornes A, Heymons N, Bange G, Schuldiner M, Zalckvar E, Bölker M, Schekman R, Freitag J. Bittner E, et al. PLoS Biol. 2024 Feb 20;22(2):e3002508. doi: 10.1371/journal.pbio.3002508. eCollection 2024 Feb. PLoS Biol. 2024. PMID: 38377076 Free PMC article. - Mitochondria-Associated ER Membranes - The Origin Site of Autophagy.
Yang M, Li C, Yang S, Xiao Y, Xiong X, Chen W, Zhao H, Zhang Q, Han Y, Sun L. Yang M, et al. Front Cell Dev Biol. 2020 Jul 16;8:595. doi: 10.3389/fcell.2020.00595. eCollection 2020. Front Cell Dev Biol. 2020. PMID: 32766245 Free PMC article. Review. - Mitochondrial association, protein phosphorylation, and degradation regulate the availability of the active Rab GTPase Ypt11 for mitochondrial inheritance.
Lewandowska A, Macfarlane J, Shaw JM. Lewandowska A, et al. Mol Biol Cell. 2013 Apr;24(8):1185-95. doi: 10.1091/mbc.E12-12-0848. Epub 2013 Feb 20. Mol Biol Cell. 2013. PMID: 23427260 Free PMC article. - Means of intracellular communication: touching, kissing, fusing.
Spang A. Spang A. Microb Cell. 2021 Apr 13;8(5):87-90. doi: 10.15698/mic2021.05.747. Microb Cell. 2021. PMID: 33981760 Free PMC article.
References
- Voelker DR. J Lipid Res. 2003;44:441. - PubMed
- Achleitner G, et al. Eur J Biochem. 1999;264:545. - PubMed
- Vance JE. J Biol Chem. 1991;266:89. - PubMed
- Rizzuto R, Simpson AW, Brini M, Pozzan T. Nature. 1992;358:325. - PubMed
- Rizzuto R, Brini M, Murgia M, Pozzan T. Science. 1993;262:744. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases