Identifying relationships among genomic disease regions: predicting genes at pathogenic SNP associations and rare deletions - PubMed (original) (raw)
Identifying relationships among genomic disease regions: predicting genes at pathogenic SNP associations and rare deletions
Soumya Raychaudhuri et al. PLoS Genet. 2009 Jun.
Abstract
Translating a set of disease regions into insight about pathogenic mechanisms requires not only the ability to identify the key disease genes within them, but also the biological relationships among those key genes. Here we describe a statistical method, Gene Relationships Among Implicated Loci (GRAIL), that takes a list of disease regions and automatically assesses the degree of relatedness of implicated genes using 250,000 PubMed abstracts. We first evaluated GRAIL by assessing its ability to identify subsets of highly related genes in common pathways from validated lipid and height SNP associations from recent genome-wide studies. We then tested GRAIL, by assessing its ability to separate true disease regions from many false positive disease regions in two separate practical applications in human genetics. First, we took 74 nominally associated Crohn's disease SNPs and applied GRAIL to identify a subset of 13 SNPs with highly related genes. Of these, ten convincingly validated in follow-up genotyping; genotyping results for the remaining three were inconclusive. Next, we applied GRAIL to 165 rare deletion events seen in schizophrenia cases (less than one-third of which are contributing to disease risk). We demonstrate that GRAIL is able to identify a subset of 16 deletions containing highly related genes; many of these genes are expressed in the central nervous system and play a role in neuronal synapses. GRAIL offers a statistically robust approach to identifying functionally related genes from across multiple disease regions--that likely represent key disease pathways. An online version of this method is available for public use (http://www.broad.mit.edu/mpg/grail/).
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
Figure 1. Gene Relationships Among Implicated Loci (GRAIL) method consists of four steps.
(A) Identifying genes in disease regions. For each independent associated SNP or CNV from a GWA study, GRAIL defines a disease region; then GRAIL identifies genes overlapping the region. In this region there are three genes. We use gene 1 (pink arrow) as an example. (B) Assess relatedness to other human genes. GRAIL scores each gene contained in a disease region for relatedness to all other human genes. GRAIL determines gene relatedness by looking at words in gene references; related genes are defined as those whose abstract references use similar words. Here gene 1 has word counts that are highly similar to gene A but not to gene B. All human genes are ranked according to text-based similarity (green bar), and the most similar genes are considered related. (C) Counting regions with similar genes. For each gene in a disease region, GRAIL assesses whether other independent disease regions contain highly significant genes. GRAIL assigns a significance score to the count. In this illustration gene 1 is similar to genes in three of the regions (green arrows), including gene A. (D) Assigning a significance score to a disease region. After all of the genes within a region are scored, GRAIL identifies the most significant gene as the likely candidate. GRAIL corrects its significance score for multiple hypothesis testing (by adjusting for the number of genes in the region), to assign a significance score to the region.
Figure 2. SNPs associated with lipid metabolism and height contain genes related to each other.
(A) 19 SNPs associated with lipid metabolism. The _y_-axis plots the ptext values on a log scale, with increasing significance at the top. The histogram on the left side of the graph illustrates values for matched SNP sets. 88.6% of those SNPs have ptext values that are >0.1. The scatter plot on the right illustrates ptext values for actual serum cholesterol associated SNPs (blue dots). Black horizontal line marks the median ptext value. We assessed the same SNP with similarity metrics based on gene annotation (green dots) and gene expression correlation (purple dots). (B) 42 SNPs associated with height. Similar plot for 42 height associated SNPs. The histogram on the left of the graph illustrates ptext values for random SNP sets carefully matched to height-associated SNP set. 86.5% of those SNPs have ptext values that are >0.1. The scatter plot on the right illustrates ptext values for actual SNPs associated with height (blue dots). Black horizontal line marks the median ptext value. We assessed the same SNP with similarity metrics based on gene annotation (green dots) and gene expression correlation (purple dots). On the right we list for each ptext threshold the number of expected SNPs less than the threshold based on matched sets, and the number of observed SNPs less than the threshold among height associated SNPs.
Figure 3. GRAIL predicts Crohn's disease SNPs.
(A) Validated versus Failed SNPs. Prior to replication, GRAIL scored Crohn's SNPs that emerged from a meta-analysis study. Results from follow-up testing either validated Crohn's SNPs, or identified those SNPs that failed. We produce a scatter plot of the significance of text-based similiarty (ptext) for validated regions (green) versus regions that failed to replicate (red). Black horizontal lines mark the median ptext values. The distribution of scores for failed SNPs resembles a random distribution of _p_-values. The distribution of scores for validated SNPs is significantly different; almost ½ of these SNPs obtain ptext scores<0.1. (B) Histogram of text-based scores for Crohn's disease candidate regions. Here we plot a histogram of _p_ text scores for 74 Crohn's disease SNPs. Validated SNPs (green) have _p_ text values that are enriched for significant values. Indeterminate SNPs (yellow) have a subset of _p_ text values that are significant. Failed SNPs (Red) have all of their _p_ text scores>0.1.
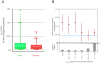
Figure 4. GRAIL identifies a subset of highly connected genes within rare deletions found in Schizophrenia cases.
(A) Case deletions versus control deletions. Here we plot the results of the separate GRAIL analyses conducted on the deletions observed in schizophrenia cases and controls. Case deletion ptext scores are displayed in red; control deletion ptext scores are displayed in green. The line in each category in the middle of the box represents the median GRAIL ptext score. The box represents the 25–75% range. The bars represent the 5–95% range. Additional scores outside the range are individual plotted. (B) Text-based GRAIL significance score tracks with CNS specific expression. We partition case-only deletions by their GRAIL scores. For each range of GRAIL ptext scores, we assess the candidate genes selected by GRAIL for CNS expression. The upper portion of this plot illustrates the fraction of those candidate genes that demonstrate preferential CNS expression along with 95% confidence intervals. The blue line represents the total fraction of genes that are preferentially CNS expressed. For the most compelling GRAIL scores, the candidate genes are significantly enriched for CNS expression compared to what would be expected from a random group of genes. The lower portion of the plot is a histogram.
Similar articles
- GenoWAP: GWAS signal prioritization through integrated analysis of genomic functional annotation.
Lu Q, Yao X, Hu Y, Zhao H. Lu Q, et al. Bioinformatics. 2016 Feb 15;32(4):542-8. doi: 10.1093/bioinformatics/btv610. Epub 2015 Oct 25. Bioinformatics. 2016. PMID: 26504140 Free PMC article. - VIZ-GRAIL: visualizing functional connections across disease loci.
Raychaudhuri S. Raychaudhuri S. Bioinformatics. 2011 Jun 1;27(11):1589-90. doi: 10.1093/bioinformatics/btr185. Epub 2011 Apr 19. Bioinformatics. 2011. PMID: 21505031 Free PMC article. - Rare CNVs in Suicide Attempt include Schizophrenia-Associated Loci and Neurodevelopmental Genes: A Pilot Genome-Wide and Family-Based Study.
Sokolowski M, Wasserman J, Wasserman D. Sokolowski M, et al. PLoS One. 2016 Dec 28;11(12):e0168531. doi: 10.1371/journal.pone.0168531. eCollection 2016. PLoS One. 2016. PMID: 28030616 Free PMC article. - Predicting novel genomic regions linked to genetic disorders using GWAS and chromosome conformation data - a case study of schizophrenia.
Buxton DS, Batten DJ, Crofts JJ, Chuzhanova N. Buxton DS, et al. Sci Rep. 2019 Nov 29;9(1):17940. doi: 10.1038/s41598-019-54514-2. Sci Rep. 2019. PMID: 31784692 Free PMC article. - Classification of genetic profiles of Crohn's disease: a focus on the ATG16L1 gene.
Grant SF, Baldassano RN, Hakonarson H. Grant SF, et al. Expert Rev Mol Diagn. 2008 Mar;8(2):199-207. doi: 10.1586/14737159.8.2.199. Expert Rev Mol Diagn. 2008. PMID: 18366306 Review.
Cited by
- Unravelling the Oral-Gut Axis: Interconnection Between Periodontitis and Inflammatory Bowel Disease, Current Challenges, and Future Perspective.
Tanwar H, Gnanasekaran JM, Allison D, Chuang LS, He X, Aimetti M, Baima G, Costalonga M, Cross RK, Sears C, Mehandru S, Cho J, Colombel JF, Raufman JP, Thumbigere-Math V. Tanwar H, et al. J Crohns Colitis. 2024 Aug 14;18(8):1319-1341. doi: 10.1093/ecco-jcc/jjae028. J Crohns Colitis. 2024. PMID: 38417137 Free PMC article. Review. - SNRPD1 conveys prognostic value on breast cancer survival and is required for anthracycline sensitivity.
Dai X, Cai L, Zhang Z, Li J. Dai X, et al. BMC Cancer. 2023 Apr 25;23(1):376. doi: 10.1186/s12885-023-10860-z. BMC Cancer. 2023. PMID: 37098488 Free PMC article. - A unifying statistical framework to discover disease genes from GWASs.
McManus JNJ, Lovelett RJ, Lowengrub D, Christensen S. McManus JNJ, et al. Cell Genom. 2023 Mar 8;3(3):100264. doi: 10.1016/j.xgen.2023.100264. eCollection 2023 Mar 8. Cell Genom. 2023. PMID: 36950381 Free PMC article. - Network expansion of genetic associations defines a pleiotropy map of human cell biology.
Barrio-Hernandez I, Schwartzentruber J, Shrivastava A, Del-Toro N, Gonzalez A, Zhang Q, Mountjoy E, Suveges D, Ochoa D, Ghoussaini M, Bradley G, Hermjakob H, Orchard S, Dunham I, Anderson CA, Porras P, Beltrao P. Barrio-Hernandez I, et al. Nat Genet. 2023 Mar;55(3):389-398. doi: 10.1038/s41588-023-01327-9. Epub 2023 Feb 23. Nat Genet. 2023. PMID: 36823319 Free PMC article.
References
- Gudbjartsson DF, Walters GB, Thorleifsson G, Stefansson H, Halldorsson BV, et al. Many sequence variants affecting diversity of adult human height. Nat Genet. 2008;40:609–615. - PubMed
Publication types
MeSH terms
Grants and funding
- P30 DK040561/DK/NIDDK NIH HHS/United States
- U01 HG004171/HG/NHGRI NIH HHS/United States
- T32 GM007753/GM/NIGMS NIH HHS/United States
- T32 AR007530/AR/NIAMS NIH HHS/United States
- U01HG004171/HG/NHGRI NIH HHS/United States
- T32AR007530-23/AR/NIAMS NIH HHS/United States
- K08 AR055688/AR/NIAMS NIH HHS/United States
- K08 AR055688-01A1/AR/NIAMS NIH HHS/United States
- 1K08AR055688-01A1/AR/NIAMS NIH HHS/United States
- P30 DK040561-14/DK/NIDDK NIH HHS/United States
- R01 DK083759/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical