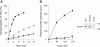Loss-of-function mutation in the dioxygenase-encoding FTO gene causes severe growth retardation and multiple malformations - PubMed (original) (raw)
doi: 10.1016/j.ajhg.2009.06.002. Epub 2009 Jun 25.
Orit Reish, Karine Proulx, Hiroko Kawagoe-Takaki, Barbara Sedgwick, Giles S H Yeo, David Meyre, Christelle Golzio, Florence Molinari, Noman Kadhom, Heather C Etchevers, Vladimir Saudek, I Sadaf Farooqi, Philippe Froguel, Tomas Lindahl, Stephen O'Rahilly, Arnold Munnich, Laurence Colleaux
Affiliations
- PMID: 19559399
- PMCID: PMC2706958
- DOI: 10.1016/j.ajhg.2009.06.002
Loss-of-function mutation in the dioxygenase-encoding FTO gene causes severe growth retardation and multiple malformations
Sarah Boissel et al. Am J Hum Genet. 2009 Jul.
Abstract
FTO is a nuclear protein belonging to the AlkB-related non-haem iron- and 2-oxoglutarate-dependent dioxygenase family. Although polymorphisms within the first intron of the FTO gene have been associated with obesity, the physiological role of FTO remains unknown. Here we show that a R316Q mutation, inactivating FTO enzymatic activity, is responsible for an autosomal-recessive lethal syndrome. Cultured skin fibroblasts from affected subjects showed impaired proliferation and accelerated senescence. These findings indicate that FTO is essential for normal development of the central nervous and cardiovascular systems in human and establish that a mutation in a human member of the AlkB-related dioxygenase family results in a severe polymalformation syndrome.
Figures
Figure 1
Genetic Analysis of a Family with Ante- and Postnatal Growth Retardation and a Severe Polymalformative Syndrome (A) Pedigree of the family. Filled symbols and slashes indicate affected and deceased infants. (B) Electrophoregrams showing the variation of FTO gene sequence in an affected infant and a healthy control. (C) Part of the multiple sequence alignment of FTO representative orthologs (H.s, Homo sapiens; Ma.m, Macaca mulatta; R.n, Rattus norvegicus; Mu.m, Mus musculus; C.f, Canis familiaris; E.c, Equus caballus; Or.a, Ornithorhynchus anatinus; Ov.a, Ovis aries; B.t, Bos taurus; M.d, Monodelphis domestica; G.g, Gallus gallus; D.r, Danio rerio; X.t, Xenopus tropicalis; ABH2 H.s, human ABH2; ABH3 H.s, Human ABH3; AlkB E.c, E. coli AlkB). The conserved residues are highlighted and the amino acid highlighted in red is the absolutely conserved arginine involved in the R316Q mutation in patients (Ai, affected infant). Blue strands labeled with roman numerals identify three of the eight β strands that form the conserved double-stranded β-helix of the 2OG-oxygenases.
Figure 2
Biochemical Analyses of the Wild-Type and Mutant FTO Proteins (A) Purified FTO was added to 10 μl reaction mixture containing 50 mM HEPES.KOH (pH 7), 50 μg/ml BSA, 4 mM ascorbate, 75 μM Fe(NH4)2(SO4)2, and 20 μM [5-14C]-2-oxoglutarate (30 mCi/mmol from Moravek Biochemicals) and incubated at 37°C for various times. To measure stimulation of this activity by 3-methylthymidine, 1 mM 3-methylthymidine (Moravek Biochemicals) was included in the assay mix. The reaction was stopped by adding 5 μl stop solution containing 20 mM succinate, 20 mM 2-oxoglutarate followed by 5 μl 160 mM dinitrophenylhydrazine, which precipitates 2-oxoglutarate. This mix was incubated at room temperature for 30 min. An additional 10 μl 1M 2-oxoglutarate was added and incubated for a further 30 min. The precipitate was removed by centrifugation. Clear supernatant (10 μl) was scintillation counted to monitor the 14C-succinate generated. Time course of activity of 1.5 μM FTO: open and closed squares, wild-type FTO protein; open and closed triangles, mutant R316Q FTO protein. Open symbols and dotted line: without 3-methylthymidine; closed symbols and solid line: with 3-methylthymidine (1 mM). (B) A DNA substrate containing 14C-3-methylthymine was prepared by treating poly(dT) with [14C]-methyl iodide (54 Ci/mmole, Amersham Biosciences) as previously described and had a specific activity of 1580 cpm/μg poly(dT). FTO was added to the 14C-methylated poly(dT) substrate (1000 cpm) in a 100 μl reaction mix containing 50 mM MES-HCl (pH 6), 75 μM Fe(NH4)2(SO4)2, 100 μM 2-oxoglutarate, 2 mM ascorbate, 10 μg/ml bovine serum albumin and incubated at 20°C for various times. All assays were performed in triplicate. To stop the reaction, EDTA was added to a final concentration of 10 mM. The polynucleotide substrate was then ethanol precipitated in the presence of carrier calf thymus DNA. Two-thirds of the ethanol-soluble radioactive material was monitored by scintillation counting. Equal volumes of the protein preparations were assayed. Standard error of the mean is shown for each time point. Closed square, 2 μM wild-type FTO protein; closed triangle, 2 μM mutant R316Q FTO protein; open circle, “mock prep” prepared in the absence of recombinant FTO expression. Equal volumes of the protein preparations were also examined by SDS-10% PAGE.
Figure 3
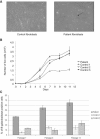
Cell Morphology and Proliferative Abilities of Cultured Skin Fibroblasts Harboring the R316Q FTO Mutation (A) Fibroblast morphology. The arrow shows altered cell morphology and enlarged cell size of a patient fibroblast. (B) Fibroblast growth curves. Fibroblasts were seeded at a density of 5000 cells/well in 12-well dishes. The number of cells per dish was determined with a CASY cell counter on days 2, 5, 7, 9, and 11 after seeding. Filled and open symbols correspond to patient and control fibroblasts, respectively. Data correspond to the mean of three replicates. Standard error of the mean is shown for each point. (C) Senescence-associated β-galactosidase assay. Fibroblasts were seeded on 6-well dishes at a density of 130,000 cells/well. Senescent fibroblasts were stained with the Senescence β-Galactosidase Staining Kit (Cell Signaling Technology). Percentages of β-galactosidase-positive cells for passages 7, 9, and 13 were calculated on the basis of approximately 100 cells. Data correspond to the mean of three replicates. Standard error of the mean is shown for each point.
Similar articles
- Identification of a pathogenic FTO mutation by next-generation sequencing in a newborn with growth retardation and developmental delay.
Daoud H, Zhang D, McMurray F, Yu A, Luco SM, Vanstone J, Jarinova O, Carson N, Wickens J, Shishodia S, Choi H, McDonough MA, Schofield CJ, Harper ME, Dyment DA, Armour CM. Daoud H, et al. J Med Genet. 2016 Mar;53(3):200-7. doi: 10.1136/jmedgenet-2015-103399. Epub 2015 Sep 16. J Med Genet. 2016. PMID: 26378117 - FTO variant associated with malformation syndrome.
Rohena L, Lawson M, Guzman E, Ganapathi M, Cho MT, Haverfield E, Anyane-Yeboa K. Rohena L, et al. Am J Med Genet A. 2016 Apr;170A(4):1023-8. doi: 10.1002/ajmg.a.37515. Epub 2015 Dec 24. Am J Med Genet A. 2016. PMID: 26697951 - The fat mass and obesity associated gene FTO functions in the brain to regulate postnatal growth in mice.
Gao X, Shin YH, Li M, Wang F, Tong Q, Zhang P. Gao X, et al. PLoS One. 2010 Nov 16;5(11):e14005. doi: 10.1371/journal.pone.0014005. PLoS One. 2010. PMID: 21103374 Free PMC article. - The biology of FTO: from nucleic acid demethylase to amino acid sensor.
Gulati P, Yeo GS. Gulati P, et al. Diabetologia. 2013 Oct;56(10):2113-21. doi: 10.1007/s00125-013-2999-5. Epub 2013 Jul 30. Diabetologia. 2013. PMID: 23896822 Free PMC article. Review. - A loop matters for FTO substrate selection.
Han Z, Huang N, Niu T, Chai J. Han Z, et al. Protein Cell. 2010 Jul;1(7):616-20. doi: 10.1007/s13238-010-0082-2. Epub 2010 Jul 29. Protein Cell. 2010. PMID: 21203933 Free PMC article. Review.
Cited by
- ALKBHs-facilitated RNA modifications and de-modifications.
A Alemu E, He C, Klungland A. A Alemu E, et al. DNA Repair (Amst). 2016 Aug;44:87-91. doi: 10.1016/j.dnarep.2016.05.026. Epub 2016 May 17. DNA Repair (Amst). 2016. PMID: 27237585 Free PMC article. Review. - Epigenetic role of N6-methyladenosine (m6A) RNA methylation in the cardiovascular system.
Zhao K, Yang CX, Li P, Sun W, Kong XQ. Zhao K, et al. J Zhejiang Univ Sci B. 2020 Jul;21(7):509-523. doi: 10.1631/jzus.B1900680. J Zhejiang Univ Sci B. 2020. PMID: 32633106 Free PMC article. Review. - Long-term exercise training down-regulates m6A RNA demethylase FTO expression in the hippocampus and hypothalamus: an effective intervention for epigenetic modification.
Liu SJ, Cai TH, Fang CL, Lin SZ, Yang WQ, Wei Y, Zhou F, Liu L, Luo Y, Guo ZY, Zhao G, Li YP, Li LM. Liu SJ, et al. BMC Neurosci. 2022 Sep 26;23(1):54. doi: 10.1186/s12868-022-00742-8. BMC Neurosci. 2022. PMID: 36163017 Free PMC article. - Association of FTO Mutations with Risk and Survival of Breast Cancer in a Chinese Population.
Zeng X, Ban Z, Cao J, Zhang W, Chu T, Lei D, Du Y. Zeng X, et al. Dis Markers. 2015;2015:101032. doi: 10.1155/2015/101032. Epub 2015 Jun 4. Dis Markers. 2015. PMID: 26146447 Free PMC article. - DNA repair by reversal of DNA damage.
Yi C, He C. Yi C, et al. Cold Spring Harb Perspect Biol. 2013 Jan 1;5(1):a012575. doi: 10.1101/cshperspect.a012575. Cold Spring Harb Perspect Biol. 2013. PMID: 23284047 Free PMC article. Review.
References
- Peters T., Ausmeier K., Ruther U. Cloning of Fatso (Fto), a novel gene deleted by the Fused toes (Ft) mouse mutation. Mamm. Genome. 1999;10:983–986. - PubMed
- Gotz K., Briscoe J., Ruther U. Homozygous Ft embryos are affected in floor plate maintenance and ventral neural tube patterning. Dev. Dyn. 2005;233:623–630. - PubMed
- Anselme I., Laclef C., Lanaud M., Ruther U., Schneider-Maunoury S. Defects in brain patterning and head morphogenesis in the mouse mutant Fused toes. Dev. Biol. 2007;304:208–220. - PubMed
- Dina C., Meyre D., Gallina S., Durand E., Korner A., Jacobson P., Carlsson L.M., Kiess W., Vatin V., Lecoeur C. Variation in FTO contributes to childhood obesity and severe adult obesity. Nat. Genet. 2007;39:724–726. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases