Absolute metabolite concentrations and implied enzyme active site occupancy in Escherichia coli - PubMed (original) (raw)
Absolute metabolite concentrations and implied enzyme active site occupancy in Escherichia coli
Bryson D Bennett et al. Nat Chem Biol. 2009 Aug.
Abstract
Absolute metabolite concentrations are critical to a quantitative understanding of cellular metabolism, as concentrations impact both the free energies and rates of metabolic reactions. Here we use LC-MS/MS to quantify more than 100 metabolite concentrations in aerobic, exponentially growing Escherichia coli with glucose, glycerol or acetate as the carbon source. The total observed intracellular metabolite pool was approximately 300 mM. A small number of metabolites dominate the metabolome on a molar basis, with glutamate being the most abundant. Metabolite concentration exceeds K(m) for most substrate-enzyme pairs. An exception is lower glycolysis, where concentrations of intermediates are near the K(m) of their consuming enzymes and all reactions are near equilibrium. This may facilitate efficient flux reversibility given thermodynamic and osmotic constraints. The data and analyses presented here highlight the ability to identify organizing metabolic principles from systems-level absolute metabolite concentration data.
Figures
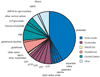
Figure 1. Composition of the measured metabolome
The pie graph shows the molar abundance of different metabolites in glucose-fed cells. Amino acids are shown in dark blue, nucleotides in rust, NAD(P)(H) in yellow, glutathiones in pink, central carbon metabolites in dark green, and all other metabolites in light blue. Abundant metabolites are labeled. Abrevations used: ATP, adenosine-5'-triphosphate; UTP, uridine-5'-triphosphate; GTP, guanosine 5'-triphosphate; dTTP, thymidine 5'-triphosphate; CTP, cytidine-5'-triphosphate; NAD+, nicotinamide adenine dinucleotide; FBP, fructose-1,6-bisphosphate; 6-P-gluconate, 6-phospho-gluconate; Hexose-P, the combined pools of glucose-6-phosphate, glucose-1-phosphate, and fructose-6-phosphate; UDP-N-Ac-Glucosamine, uridine-5'-diphosphate N-acetyl-glucosamine; UDPG, uridine-5'-diphosphate glucose.
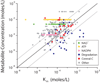
Figure 2. Implied enzyme active site saturation
The relationship of metabolite concentration and Km of their consuming enzymes in glucose-grown E. coli. NAD+ is shown as green squares, ATP as yellow squares, NADPH as pinksquares, degradation reactions as blue circles, and reactions in central carbon metabolism (glycolysis, the pentose-phosphate pathway, and the TCA cycle) as orange circles. All other data are shown as grey diamonds. The dark line is the line of unity (where concentration = Km) and the light lines denote a 10-fold deviation from the line of unity.
Figure 3
Comment in
- A metabolic network described in absolute terms.
Schomburg D. Schomburg D. Nat Chem Biol. 2009 Aug;5(8):535-6. doi: 10.1038/nchembio0809-535. Nat Chem Biol. 2009. PMID: 19620991 No abstract available.
Similar articles
- Metabolite concentrations, fluxes and free energies imply efficient enzyme usage.
Park JO, Rubin SA, Xu YF, Amador-Noguez D, Fan J, Shlomi T, Rabinowitz JD. Park JO, et al. Nat Chem Biol. 2016 Jul;12(7):482-9. doi: 10.1038/nchembio.2077. Epub 2016 May 2. Nat Chem Biol. 2016. PMID: 27159581 Free PMC article. - In vivo dynamics of glycolysis in Escherichia coli shows need for growth-rate dependent metabolome analysis.
Schaub J, Reuss M. Schaub J, et al. Biotechnol Prog. 2008 Nov-Dec;24(6):1402-7. doi: 10.1002/btpr.59. Biotechnol Prog. 2008. PMID: 19194955 - Translational Metabolomics of Head Injury: Exploring Dysfunctional Cerebral Metabolism with Ex Vivo NMR Spectroscopy-Based Metabolite Quantification.
Wolahan SM, Hirt D, Glenn TC. Wolahan SM, et al. In: Kobeissy FH, editor. Brain Neurotrauma: Molecular, Neuropsychological, and Rehabilitation Aspects. Boca Raton (FL): CRC Press/Taylor & Francis; 2015. Chapter 25. In: Kobeissy FH, editor. Brain Neurotrauma: Molecular, Neuropsychological, and Rehabilitation Aspects. Boca Raton (FL): CRC Press/Taylor & Francis; 2015. Chapter 25. PMID: 26269925 Free Books & Documents. Review. - Towards a systems level understanding of the oxygen response of Escherichia coli.
Bettenbrock K, Bai H, Ederer M, Green J, Hellingwerf KJ, Holcombe M, Kunz S, Rolfe MD, Sanguinetti G, Sawodny O, Sharma P, Steinsiek S, Poole RK. Bettenbrock K, et al. Adv Microb Physiol. 2014;64:65-114. doi: 10.1016/B978-0-12-800143-1.00002-6. Adv Microb Physiol. 2014. PMID: 24797925 Review.
Cited by
- Oxidation of fatty aldehydes to fatty acids by Escherichia coli cells expressing the Vibrio harveyi fatty aldehyde dehydrogenase (FALDH).
Buchhaupt M, Guder J, Sporleder F, Paetzold M, Schrader J. Buchhaupt M, et al. World J Microbiol Biotechnol. 2013 Mar;29(3):569-75. doi: 10.1007/s11274-012-1211-2. Epub 2012 Nov 21. World J Microbiol Biotechnol. 2013. PMID: 23180547 - Underground isoleucine biosynthesis pathways in E. coli.
Cotton CA, Bernhardsgrütter I, He H, Burgener S, Schulz L, Paczia N, Dronsella B, Erban A, Toman S, Dempfle M, De Maria A, Kopka J, Lindner SN, Erb TJ, Bar-Even A. Cotton CA, et al. Elife. 2020 Aug 24;9:e54207. doi: 10.7554/eLife.54207. Elife. 2020. PMID: 32831171 Free PMC article. - Pyrophosphate-Dependent ATP Formation from Acetyl Coenzyme A in Syntrophus aciditrophicus, a New Twist on ATP Formation.
James KL, Ríos-Hernández LA, Wofford NQ, Mouttaki H, Sieber JR, Sheik CS, Nguyen HH, Yang Y, Xie Y, Erde J, Rohlin L, Karr EA, Loo JA, Ogorzalek Loo RR, Hurst GB, Gunsalus RP, Szweda LI, McInerney MJ. James KL, et al. mBio. 2016 Aug 16;7(4):e01208-16. doi: 10.1128/mBio.01208-16. mBio. 2016. PMID: 27531911 Free PMC article. - Cofactor Specificity of Glucose-6-Phosphate Dehydrogenase Isozymes in Pseudomonas putida Reveals a General Principle Underlying Glycolytic Strategies in Bacteria.
Volke DC, Olavarría K, Nikel PI. Volke DC, et al. mSystems. 2021 Mar 16;6(2):e00014-21. doi: 10.1128/mSystems.00014-21. mSystems. 2021. PMID: 33727391 Free PMC article. - Increasing the Thermodynamic Driving Force of the Phosphofructokinase Reaction in Clostridium thermocellum.
Hon S, Jacobson T, Stevenson DM, Maloney MI, Giannone RJ, Hettich RL, Amador-Noguez D, Olson DG, Lynd LR. Hon S, et al. Appl Environ Microbiol. 2022 Nov 22;88(22):e0125822. doi: 10.1128/aem.01258-22. Epub 2022 Oct 26. Appl Environ Microbiol. 2022. PMID: 36286488 Free PMC article.
References
- Bajad SU, et al. Separation and quantitation of water soluble cellular metabolites by hydrophilic interaction chromatography-tandem mass spectrometry. J Chromatogr A. 2006;1125:76–88. - PubMed
- Coulier L, et al. Simultaneous quantitative analysis of metabolites using ion-pair liquid chromatography-electrospray ionization mass spectrometry. Anal Chem. 2006;78:6573–6582. - PubMed
- Luo B, Groenke K, Takors R, Wandrey C, Oldiges M. Simultaneous determination of multiple intracellular metabolites in glycolysis, pentose phosphate pathway and tricarboxylic acid cycle by liquid chromatography-mass spectrometry. J Chromatogr A. 2007;1147:153–164. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases