Remote homology between Munc13 MUN domain and vesicle tethering complexes - PubMed (original) (raw)
Remote homology between Munc13 MUN domain and vesicle tethering complexes
Jimin Pei et al. J Mol Biol. 2009.
Abstract
Most core components of the neurotransmitter release machinery have homologues in other types of intracellular membrane traffic, likely underlying a universal mechanism of intracellular membrane fusion. However, no clear similarity between Munc13s and protein families generally involved in membrane traffic has been reported, despite the essential nature of Munc13s for neurotransmitter release. This crucial function was ascribed to a minimal Munc13 region called the MUN domain, which likely participates in soluble N-ethylmaleimide sensitive factor attachment protein receptor complex (SNARE) assembly and is also found in Ca(2+)-dependent activator protein for secretion. We have now used comparative sequence and structural analyses to study the structure and evolutionary origin of the MUN domain. We found weak yet significant sequence similarities between the MUN domain and a set of protein subunits from several related vesicle tethering complexes, such as Sec6 from the exocyst complex and Vps53 from the Golgi-associated retrograde protein complex. Such an evolutionary relationship allows structure prediction of the MUN domain and suggests functional similarities between MUN domain-containing proteins and multisubunit tethering complexes such as exocyst, conserved oligomeric Golgi complex, Golgi-associated retrograde protein complex, and Dsl1p. These findings further unify the mechanism of neurotransmitter release with those of other types of intracellular membrane traffic and, in turn, support a role for tethering complexes in soluble N-ethylmaleimide sensitive factor attachment protein receptor complex assembly.
Figures
Figure 1. Multiple sequence alignment of domain C and domain D of the MUN domains
This alignment was constructed by the PROMALS3D program with manual adjustment according to available 3D structures and secondary structure predictions. The proteins are identified by their NCBI gene identification (gi) numbers. Their grouping is suggested by boxes around the gi numbers. Common names for some experimentally characterized proteins follow the gi numbers. PDB ids for sequences with known structures are shaded and shown after the common names. Non-polar residues in positions with mainly hydrophobic residues are shaded in yellow. Conserved residues in the MUN domain-containing proteins are shown in red and bold letters, and corresponding residues in proteins from vesicle tethering complexes are shown in black and bold letters. Starting and ending residues numbers (italic), as well as sequence lengths (in brackets), are shown. Insertion regions between the aligned blocks are replaced by the numbers of residues. The long insertion regions with the C2 domains in fungi MUN domain-containing proteins are indicated. Consensus secondary structure predictions for the MUN domains are shown above the alignment and marked from h1 to h9 for the nine major helices in domain C and domain D of tethering complex subunits such as Exo70 and Tip20p. Species names are represented by two letter abbreviations and colored as follows: metazoan: blue; fungi: orange; and plants: green. Species name abbreviations are: At: Arabidopsis thaliana; Ce: Caenorhabditis elegans; Ci: Ciona intestinalis; Dm: Drosophila melanogaster; Mm: Mus musculus; Nc: Neurospora crassa; Ot: Ostreococcus tauri; Sc: Saccharomyces cerevisiae; Sp: Schizosaccharomyces pombe.
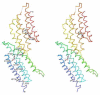
Figure 2. Stereo diagrams of the structural model of domain C and domain D of the MUN domain from mouse Munc13-1
α-Helices are labeled in accordance with Figure 1. Sidechains of conserved residues (highlighted in red letters in Figure 1) are shown as sticks. N- and C-termini are marked. Long loops are omitted with dotted lines connecting the end points. These diagrams are made by program PyMOL.
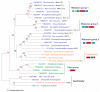
Figure 3. Phylogenetic tree of representative MUN domains and Sec6 proteins (as an out group) by MOLPHY
Sequence groups are marked on the right with domain architecture diagrams shown. The dashed outline of the first C2 domain in metazoan group 1 suggests that it is absent in some proteins. JTT amino acid substitution model was used. Only positions with gap fraction less than 50% were selected for tree building. The local estimates of bootstrap percentages were obtained by the RELL method, as implemented in the program ProtML of MOLPHY (-R option). Branch support values above 90 are shown in red numbers. A similar tree supporting the separation of five major groups of MUN domains were obtained by using the PHYML program with the JTT amino acid substitution model and with rate variation across sites modeled by a discrete gamma-distribution.
Similar articles
- The crystal structure of a Munc13 C-terminal module exhibits a remarkable similarity to vesicle tethering factors.
Li W, Ma C, Guan R, Xu Y, Tomchick DR, Rizo J. Li W, et al. Structure. 2011 Oct 12;19(10):1443-55. doi: 10.1016/j.str.2011.07.012. Structure. 2011. PMID: 22000513 Free PMC article. - The Dsl1 tethering complex actively participates in soluble NSF (N-ethylmaleimide-sensitive factor) attachment protein receptor (SNARE) complex assembly at the endoplasmic reticulum in Saccharomyces cerevisiae.
Diefenbacher M, Thorsteinsdottir H, Spang A. Diefenbacher M, et al. J Biol Chem. 2011 Jul 15;286(28):25027-38. doi: 10.1074/jbc.M110.215657. Epub 2011 Apr 11. J Biol Chem. 2011. PMID: 21482823 Free PMC article. - Golgi inCOGnito: From vesicle tethering to human disease.
D'Souza Z, Taher FS, Lupashin VV. D'Souza Z, et al. Biochim Biophys Acta Gen Subj. 2020 Nov;1864(11):129694. doi: 10.1016/j.bbagen.2020.129694. Epub 2020 Jul 27. Biochim Biophys Acta Gen Subj. 2020. PMID: 32730773 Free PMC article. Review. - Tethering the assembly of SNARE complexes.
Hong W, Lev S. Hong W, et al. Trends Cell Biol. 2014 Jan;24(1):35-43. doi: 10.1016/j.tcb.2013.09.006. Epub 2013 Oct 9. Trends Cell Biol. 2014. PMID: 24119662 Review.
Cited by
- Maintaining order: COG complex controls Golgi trafficking, processing, and sorting.
Blackburn JB, D'Souza Z, Lupashin VV. Blackburn JB, et al. FEBS Lett. 2019 Sep;593(17):2466-2487. doi: 10.1002/1873-3468.13570. Epub 2019 Aug 16. FEBS Lett. 2019. PMID: 31381138 Free PMC article. Review. - Functional synergy between the Munc13 C-terminal C1 and C2 domains.
Liu X, Seven AB, Camacho M, Esser V, Xu J, Trimbuch T, Quade B, Su L, Ma C, Rosenmund C, Rizo J. Liu X, et al. Elife. 2016 May 23;5:e13696. doi: 10.7554/eLife.13696. Elife. 2016. PMID: 27213521 Free PMC article. - Localization of the Priming Factors CAPS1 and CAPS2 in Mouse Sensory Neurons Is Determined by Their N-Termini.
Staudt A, Ratai O, Bouzouina A, Fecher-Trost C, Shaaban A, Bzeih H, Horn A, Shaib AH, Klose M, Flockerzi V, Lauterbach MA, Rettig J, Becherer U. Staudt A, et al. Front Mol Neurosci. 2022 Apr 14;15:674243. doi: 10.3389/fnmol.2022.674243. eCollection 2022. Front Mol Neurosci. 2022. PMID: 35493323 Free PMC article. - BAIAP3, a C2 domain-containing Munc13 protein, controls the fate of dense-core vesicles in neuroendocrine cells.
Zhang X, Jiang S, Mitok KA, Li L, Attie AD, Martin TFJ. Zhang X, et al. J Cell Biol. 2017 Jul 3;216(7):2151-2166. doi: 10.1083/jcb.201702099. Epub 2017 Jun 16. J Cell Biol. 2017. PMID: 28626000 Free PMC article. - Sec3 promotes the initial binary t-SNARE complex assembly and membrane fusion.
Yue P, Zhang Y, Mei K, Wang S, Lesigang J, Zhu Y, Dong G, Guo W. Yue P, et al. Nat Commun. 2017 Jan 23;8:14236. doi: 10.1038/ncomms14236. Nat Commun. 2017. PMID: 28112172 Free PMC article.
References
- Jahn R, Scheller RH. SNAREs--engines for membrane fusion. Nat Rev Mol Cell Biol. 2006;7:631–43. - PubMed
- Sudhof TC. The synaptic vesicle cycle. Annu Rev Neurosci. 2004;27:509–47. - PubMed
- Augustin I, Rosenmund C, Sudhof TC, Brose N. Munc13-1 is essential for fusion competence of glutamatergic synaptic vesicles. Nature. 1999;400:457–61. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 NS037200/NS/NINDS NIH HHS/United States
- GM67165/GM/NIGMS NIH HHS/United States
- R01 NS037200-02/NS/NINDS NIH HHS/United States
- R01 GM067165-02/GM/NIGMS NIH HHS/United States
- NS37200/NS/NINDS NIH HHS/United States
- R01 GM067165/GM/NIGMS NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
LinkOut - more resources
Full Text Sources
Miscellaneous