Distinct cytoplasmic maturation steps of 40S ribosomal subunit precursors require hRio2 - PubMed (original) (raw)
Distinct cytoplasmic maturation steps of 40S ribosomal subunit precursors require hRio2
Ivo Zemp et al. J Cell Biol. 2009.
Abstract
During their biogenesis, 40S ribosomal subunit precursors are exported from the nucleus to the cytoplasm, where final maturation occurs. In this study, we show that the protein kinase human Rio2 (hRio2) is part of a late 40S preribosomal particle in human cells. Using a novel 40S biogenesis and export assay, we analyzed the contribution of hRio2 to late 40S maturation. Although hRio2 is not absolutely required for pre-40S export, deletion of its binding site for the export receptor CRM1 decelerated the kinetics of this process. Moreover, in the absence of hRio2, final cytoplasmic 40S maturation is blocked because the recycling of several trans-acting factors and cytoplasmic 18S-E precursor ribosomal RNA (rRNA [pre-rRNA]) processing are defective. Intriguingly, the physical presence of hRio2 but not its kinase activity is necessary for the release of hEnp1 from cytoplasmic 40S precursors. In contrast, hRio2 kinase activity is essential for the recycling of hDim2, hLtv1, and hNob1 as well as for 18S-E pre-rRNA processing. Thus, hRio2 is involved in late 40S maturation at several distinct steps.
Figures
Figure 1.
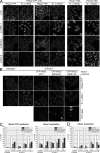
hRio2 is a component of late 40S precursors in human cells. (A) hRio2 and hEnp1 cosediment with pre-40S or 40S subunits. Cell extract was centrifuged on a 10–45% sucrose gradient. Fractions were analyzed by Western blotting. (B) hRio2 coprecipitates a pre-40S particle. hRio2 was immunoprecipitated from pooled 40S-containing gradient fractions. Proteins coprecipitated with hRio2 were analyzed by silver staining and identified by MS. (C) Western blot analysis confirms the presence of hRio2, hEnp1, hLtv1, and hRps3 in the hRio2-associated particle. (D) hRio2-associated particles contain 18S-E pre-rRNA. rRNA was isolated from the hRio2 immunoprecipitate and analyzed by Northern blotting.
Figure 2.
hRio2 is exported from the nucleus in a CRM1-dependent manner. (A) hRio2 accumulates in the nucleoplasm upon CRM1 inhibition. HeLaK cells were treated with 20 nM LMB or solvent (ethanol) for the times indicated, followed by fixation and immunofluorescence (IF) analysis for hRio2 or hEnp1. (B) Possible alignments of the NES consensus (for review see Kutay and Güttinger, 2005) to aa 391–420 of hRio2 and selected Rio2 homologues. Rio2 sequences from Homo sapiens (Hs), Mus musculus (Mm), Rattus norvegicus (Rn), Bos taurus (Bt), Canis familiaris (Cf), Xenopus laevis (Xl), and Danio rerio (Dr) were aligned using ClustalW. Φ, large, hydrophobic amino acid (underlined and bold in Rio2 sequences); x, any amino acid. Leu400 and Ile403, the two residues mutated in hRio2(NESmut), are depicted in red. (C) Mutation of the NES of hRio2 leads to its nuclear accumulation. HeLaK cells were transiently transfected with EGFP-hRio2, EGFP-hRio2(NESmut) or EGFP-hRio2(NESΔ10). 24 h after transfection, cells were analyzed by fluorescence microscopy and Western blotting for the localization and expression of hRio2 constructs, respectively. (D) The NES of hRio2 mediates CRM1-dependent nuclear export of a reporter. HeLaK cells were transiently transfected with EGFP, EGFP-NES(Nmd3), or EGFP-NES(Rio2). 24 h after transfection, cells were treated with 20 nM LMB or solvent (ethanol) for 30 min, followed by fixation and fluorescence microscopy. (E) hRio2 directly binds CRM1 in the presence of RanGTP, dependent on its NES. Recombinant zz-hRio2 WT, NESmut, and NESΔ10 were immobilized on IgG Sepharose and incubated with CRM1 in the presence or absence of hRanQ69L-GTP. Bound proteins were analyzed by Coomassie staining. (F) hRio2 WT, NESmut, and NESΔ10 display autophosphorylation activity in vitro. zz-tagged hRio2 constructs were tested for autophosphorylation activity as described in Materials and methods. Bars, 20 µm.
Figure 3.
Inducible hRps2-YFP reporter cells to study 40S subunit biogenesis. (A) Inducible hRps2-YFP reporter cells show nuclear accumulation of hRps2-YFP upon CRM1 inhibition. hRps2-YFP expression in reporter cells was induced by the addition of tet followed by treatment with 20 nM LMB or solvent (ethanol) for the times indicated. (B) Western blot analysis of A. Bar, 20 µm.
Figure 4.
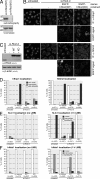
hRio2 is not absolutely required for pre-40S export but contributes to 40S export efficiency. (A) hRio2 depletion causes pre-40S export defects in hRps2-YFP reporter cells and HeLaY cells but not in HeLaK cells. Cells were transfected with the siRNAs (si) indicated. In reporter cells, hRps2-YFP expression was induced 35 h after transfection. 48 h after transfection, cells were fixed, followed by immunofluorescence (IF) analysis for hRio2 or hNob1. Note that ∼25–30% of reporter cells and 10–15% of HeLaY cells displayed nuclear accumulation of hRps2-YFP or hNob1. ctrl, control. (B) Pre-40S export defects upon hRio2 depletion can be rescued by overexpression of CFP-tagged hRio2(WT) but not hRio2(NESmut) or hRio2(NESΔ10). RNAi experiments were performed using hRps2-YFP reporter cells as in A, but, in addition, cells were transfected with CFP-tagged hRio2 constructs after 24 h of RNAi. (C) Quantification of the experiment shown in B. Cells displaying predominantly nuclear, intermediate, or predominantly cytoplasmic hRps2-YFP or hNob1 localization were counted. For rescues, hRps2-YFP or hNob1 localization was analyzed for transfected cells only. n, number of cells counted (from one representative experiment). (D) Nuclear accumulation of hNob1 in the nucleus of HeLaY cells after hRio2 depletion is rescued by overexpression of EGFP-tagged hRio2 but not hRio2(NESmut) or hRio2(NESΔ10). Rescue experiments using HeLaY cells were performed as in B and analyzed as in C. Bars, 20 µm.
Figure 5.
In the absence of hRio2, the trans-acting factors hNob1, hLtv1, hEnp1, and hDim2 accumulate on cytoplasmic 40S precursors that are stalled in their maturation. (A) hNob1, hLtv1, hEnp1, and hDim2 accumulate in the cytoplasm of hRio2-depleted cells. RNAi experiments in HeLaK cells were performed as described in Fig. 4 A, followed by immunofluorescence (IF) analysis for hNob1, hLtv1, hEnp1, and hDim2. (B) Western blot analysis of the RNAi experiment in A. (C) hNob1 and hLtv1 display slower shuttling kinetics in hRio2-depleted cells. 48 h after transfection, cells from the RNAi experiment in A were treated with 20 nM LMB for 1 or 2 h, followed by fixation and immunofluorescence analysis for hNob1 and hLtv1. (D) hNob1, hLtv1, hEnp1, and hDim2 are associated with 40S precursors in hRio2-depleted cells. Cell extracts from control and hRio2-depleted cells were centrifuged on a 10–45% sucrose gradient. Samples of total cells, cell extract, and pellet obtained during extract preparation as well as gradient fractions were analyzed by Western blotting. ctrl, control; si, siRNA. Bars, 20 µm.
Figure 6.
hRio2 kinase activity is required for the recycling of hDim2, hLtv1, and hNob1 but not hEnp1 from cytoplasmic 40S precursors. (A) Mutation of Lys123 and Asp246 of hRio2 to Ala results in a KD hRio2 mutant. zz-tagged hRio2(WT) and hRio2(K123A,D246A) (hRio2[KD]) were tested for autophosphorylation activity as described in Materials and methods. (B) In hRio2-depleted cells, KD hRio2 rescues the relocalization to the cytoplasm of hEnp1 but not hDim2. Rescue experiments in HeLaK cells were performed as described in Fig. 4 B using EGFP-hRio2(WT) and EGFP-hRio2(KD) as rescue constructs. (C) Western blot analysis of A revealing efficient hRio2 knockdown by RNAi and similar expression levels of the rescue constructs. (D) Quantification of the experiment shown in A. Cells displaying predominantly nucleolar, intermediate, or partially cytoplasmic hEnp1 or hDim2 localization were counted. For rescues, hEnp1 and hDim2 localization was analyzed for transfected cells only. (E and F) Slower shuttling kinetics of hLtv1 and hNob1 after hRio2 depletion are rescued by WT but not KD hRio2. Rescue experiments were performed as described in B and combined with treatment with 20 nM LMB for 1 h as described in Fig. 5 C. Cells were analyzed by immunostaining for hLtv1 (E) and hNob1 (F) and counted as in Fig. 4 C. For rescues, only transfected cells were counted. n, number of cells counted (from one representative experiment); si, siRNA. Bars, 20 µm.
Figure 7.
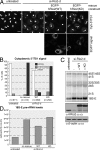
hRio2 kinase activity is required for cytoplasmic 18S-E pre-rRNA processing. (A) Cytoplasmic accumulation of 18S-E pre-rRNA after hRio2 knockdown is rescued by WT but not KD hRio2. HeLaK cells were transfected with Rio2-d siRNA (si-Rio2-d). Rescue constructs were transfected after 48 h of RNAi. Cells were fixed after 72 h of RNAi, followed by FISH analysis. FISH pictures were processed in parallel by enhancing levels, followed by setting γ correction to 0.75. Note that the 5′ ITS1 probe shows the localization of all 18S rRNA precursors, but only 18S-E pre-rRNA is detected in the cytoplasm (Rouquette et al., 2005). (B) Quantification of the experiment shown in A. Cells displaying weak, intermediate, or strong cytoplasmic 5′ ITS1 staining were counted. For rescues, only transfected cells were counted. n, number of cells counted (from one representative experiment). (C) 18S-E pre-rRNA accumulation upon hRio2 depletion is rescued by WT but not KD hRio2. Rescue experiments were performed as in A, followed by Northern blot analysis. Western blotting revealed efficient hRio2 knockdown by RNAi and similar expression levels of the rescue constructs. Note that transfection efficiencies were 39% for EGFP-hRio2(WT) and 41% for EGFP-hRio2(KD). (D) 18S-E pre-rRNA levels from the experiment in C were quantified using IQMac (GE Healthcare). Bar, 20 µm.
Similar articles
- The kinase activity of human Rio1 is required for final steps of cytoplasmic maturation of 40S subunits.
Widmann B, Wandrey F, Badertscher L, Wyler E, Pfannstiel J, Zemp I, Kutay U. Widmann B, et al. Mol Biol Cell. 2012 Jan;23(1):22-35. doi: 10.1091/mbc.E11-07-0639. Epub 2011 Nov 9. Mol Biol Cell. 2012. PMID: 22072790 Free PMC article. - Human RioK3 is a novel component of cytoplasmic pre-40S pre-ribosomal particles.
Baumas K, Soudet J, Caizergues-Ferrer M, Faubladier M, Henry Y, Mougin A. Baumas K, et al. RNA Biol. 2012 Feb;9(2):162-74. doi: 10.4161/rna.18810. Epub 2012 Feb 1. RNA Biol. 2012. PMID: 22418843 Free PMC article. - CK1δ and CK1ε are components of human 40S subunit precursors required for cytoplasmic 40S maturation.
Zemp I, Wandrey F, Rao S, Ashiono C, Wyler E, Montellese C, Kutay U. Zemp I, et al. J Cell Sci. 2014 Mar 15;127(Pt 6):1242-53. doi: 10.1242/jcs.138719. Epub 2014 Jan 14. J Cell Sci. 2014. PMID: 24424021 - Nuclear export and cytoplasmic maturation of ribosomal subunits.
Zemp I, Kutay U. Zemp I, et al. FEBS Lett. 2007 Jun 19;581(15):2783-93. doi: 10.1016/j.febslet.2007.05.013. Epub 2007 May 11. FEBS Lett. 2007. PMID: 17509569 Review. - Maturation of pre-40S particles in yeast and humans.
Cerezo E, Plisson-Chastang C, Henras AK, Lebaron S, Gleizes PE, O'Donohue MF, Romeo Y, Henry Y. Cerezo E, et al. Wiley Interdiscip Rev RNA. 2019 Jan;10(1):e1516. doi: 10.1002/wrna.1516. Epub 2018 Nov 8. Wiley Interdiscip Rev RNA. 2019. PMID: 30406965 Review.
Cited by
- The von Hippel-Lindau protein pVHL inhibits ribosome biogenesis and protein synthesis.
Zhao WT, Zhou CF, Li XB, Zhang YF, Fan L, Pelletier J, Fang J. Zhao WT, et al. J Biol Chem. 2013 Jun 7;288(23):16588-16597. doi: 10.1074/jbc.M113.455121. Epub 2013 Apr 23. J Biol Chem. 2013. PMID: 23612971 Free PMC article. - Pre-ribosomal particles from nucleoli to cytoplasm.
Kubitscheck U, Siebrasse JP. Kubitscheck U, et al. Nucleus. 2024 Dec;15(1):2373052. doi: 10.1080/19491034.2024.2373052. Epub 2024 Jun 28. Nucleus. 2024. PMID: 38940456 Free PMC article. Review. - A non-canonical mechanism for Crm1-export cargo complex assembly.
Fischer U, Schäuble N, Schütz S, Altvater M, Chang Y, Faza MB, Panse VG. Fischer U, et al. Elife. 2015 Apr 21;4:e05745. doi: 10.7554/eLife.05745. Elife. 2015. PMID: 25895666 Free PMC article. - Tandem affinity purification combined with inducible shRNA expression as a tool to study the maturation of macromolecular assemblies.
Wyler E, Zimmermann M, Widmann B, Gstaiger M, Pfannstiel J, Kutay U, Zemp I. Wyler E, et al. RNA. 2011 Jan;17(1):189-200. doi: 10.1261/rna.2325911. Epub 2010 Nov 19. RNA. 2011. PMID: 21097556 Free PMC article. - Targeting RIOK2 ATPase activity leads to decreased protein synthesis and cell death in acute myeloid leukemia.
Messling JE, Agger K, Andersen KL, Kromer K, Kuepper HM, Lund AH, Helin K. Messling JE, et al. Blood. 2022 Jan 13;139(2):245-255. doi: 10.1182/blood.2021012629. Blood. 2022. PMID: 34359076 Free PMC article.
References
- Bernad R., Engelsma D., Sanderson H., Pickersgill H., Fornerod M. 2006. Nup214-Nup88 nucleoporin subcomplex is required for CRM1-mediated 60 S preribosomal nuclear export.J. Biol. Chem. 281:19378–19386 - PubMed
- Bonazzi S., Güttinger S., Zemp I., Kutay U., Gademann K. 2007. Total synthesis, configuration, and biological evaluation of anguinomycin C.Angew. Chem. Int. Ed. Engl. 46:8707–8710 - PubMed
- Bradatsch B., Katahira J., Kowalinski E., Bange G., Yao W., Sekimoto T., Baumgartel V., Boese G., Bassler J., Wild K., et al. 2007. Arx1 functions as an unorthodox nuclear export receptor for the 60S preribosomal subunit.Mol. Cell. 27:767–779 - PubMed
- Fornerod M., Ohno M., Yoshida M., Mattaj I.W. 1997. CRM1 is an export receptor for leucine-rich nuclear export signals.Cell. 90:1051–1060 - PubMed
- Fromont-Racine M., Senger B., Saveanu C., Fasiolo F. 2003. Ribosome assembly in eukaryotes.Gene. 313:17–42 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases