The structure of the scaffold nucleoporin Nup120 reveals a new and unexpected domain architecture - PubMed (original) (raw)
The structure of the scaffold nucleoporin Nup120 reveals a new and unexpected domain architecture
Nina C Leksa et al. Structure. 2009.
Abstract
Nucleocytoplasmic transport is mediated by nuclear pore complexes (NPCs), enormous protein assemblies residing in circular openings in the nuclear envelope. The NPC is modular, with transient and stable components. The stable core is essentially built from two multiprotein complexes, the Y-shaped heptameric Nup84 complex and the Nic96 complex, arranged around an eightfold axis. We present the crystal structure of Nup120(1-757), one of the two short arms of the Y-shaped Nup84 complex. The protein adopts a compact oval shape built around a novel bipartite alpha-helical domain intimately integrated with a beta-propeller domain. The domain arrangement is substantially different from the Nup85*Seh1 complex, which forms the other short arm of the Y. With the data presented here, we establish that all three branches of the Y-shaped Nup84 complex are tightly connected by helical interactions and that the beta-propellers likely form interaction site(s) to neighboring complexes.
Figures
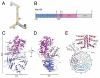
Figure 1. Overall Topology of Nup120
(A) Current model of the Y-shaped Nup84 subcomplex. The relative position of Nup120 is highlighted. (B) Schematic of full-length Nup120 from S. cerevisiae. Residues that form the β-propeller are colored blue, those that form the α-helical domain are purple, and those not present in the crystallized construct are in gray. (C, D) The overall topology of Nup120 (residues 1-757 of 1037) is shown in two views rotated by 90°. The structure is gradient-colored from blue to white to magenta from N- to C-terminus. At its N-terminus, Nup120 forms a 7-bladed β-propeller. A 4-helix bundle (α1-α4) between blades 6 and 7 packs against the remainder of the helical domain (α5-α15), composed of helices wrapping around a central hydrophobic stalk of the two long helices α11 and α12. Unstructured loops absent from the final model are shown in gray. (E) A topological diagram of the Nup120 structure is shown, illustrating the 4-helix insertion between blades 6 and 7 of the propeller as well as the two central helices of the helical domain.
Figure 2. Crystal Contacts between Two Symmetry-Related Molecules
(A) One molecule of Nup120 in blue, and its symmetry mate, related by a 2-fold rotation, in orange. The β-propellers are at opposite ends while the helical domains engage in a putative domain swap between helices α15 of both molecules. (B) Close-up of the domain-swapped region, illustrating the hydrophobic nature of helix α15 and the surrounding pocket (hydrophobic residues are shown in white). In monomeric Nup120, helix α15 likely folds under (arrow) and occupies the position taken by helix α15′ (orange) of the symmetry-related molecule in the crystal.
Figure 3. Surface Conservation and Electrostatics of Nup120
(A) Surface conservation of Nup120 is shown from three different views. To illustrate the conservation of residues on the surface of Nup120, a multiple sequence alignment sampling the phylogenetic tree of budding yeasts was generated and mapped onto the surface, colored from white (not conserved) to orange (highly conserved). The view in the middle panel corresponds to the view shown in Figure 1C. A patch of highly conserved residues is apparent on the outer face of the propeller domain of Nup120. (B) The electrostatic surface potential of Nup120 is shown in the same views as in (A) and is colored from red (−10 kT/e) to blue (+10 kT/e).
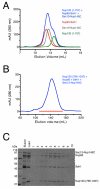
Figure 4. The C-Terminus of Nup120 Is Necessary for Binding Nup85 •Seh1 and Nup145C•Sec13
(A) Nup85•Seh1 (red), Nup145C•Sec13 (gray), and Nup1201-757 (green) were run individually and in combination (blue) on a Superdex S200 10/300 gelfiltration column. (B) Nup85•Seh1, Nup145C•Sec13, and Nup120766-1037 were incubated together and run on Superdex S200 26/60 and eluted in a single peak. (C) Fractions from the gel filtration experiment in B were analyzed by SDS-Page. Co-migration of Nup85•Seh1, Nup145C•Sec13, and Nup120766-1037 indicates that the C-terminus of Nup120 is necessary for the formation of the pentameric complex that comprises the hub of the Y-shaped complex.
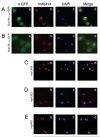
Figure 5. Nup1201-757 Does Not Localize to the Nuclear Envelope
(Aa-Ad) Nup120-GFP is targeted to the nuclear envelope, as confirmed by co-localization with mAb414 (staining FG-Nups), while Nup1201-757-GFP (Ba-Bd) is distributed throughout the cell. Mislocalization indicates that the C-terminus of Nup120 is necessary for proper recruitment to the NPC. Nup120Δ, nup133Δ, and nup84Δ cells (in the same BY4741 strain background) are shown for comparison (Cb-Cd, Db-Dd, Eb-Ed). Nuclear rim was visualized using mAb414, GFP-tagged Nup120 using goat α-GFP, and DNA using DAPI. Merged images are shown on the right.
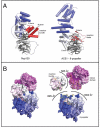
Figure 6. Nup120 is Composed of a combined β-Propeller - α-Helical Domain distinct from ACE1•β-Propeller
(A) The overall architectures of Nup120 and the ACE1 motif of Nup85•Seh1 are distinctly different. Nup120 is characterized by a bipartite helical domain (blue to white from N- to C-terminus) that is interrupted by a β-propeller domain (gray). The Nup85 ACE1 motif is characterized by an elongated helical stack (colored blue to white from N- to C-terminus) that makes a U-turn in the crown domain of the molecule. At its N-terminus, Nup85 inserts a blade (in red) into the open, 6-bladed Seh1 β-propeller. In contrast, the β-propeller of Nup120 contributes a helical insertion bundle (red) to the helical domain. The view of Nup120 is the same as that in Fig. 3B. (B) Surface representations of intact Nup120 are shown on the left, while on the right the three modules of Nup120 – the propeller, the helical insertion, and the helical domain – are shown pulled apart to illustrate the buried surface areas in between. Interacting surfaces between the propeller and the insertion bundle are outlined in green, between the propeller and the helical domain in yellow, and between the insertion bundle and the helical domain in orange. The molecule is N-to-C gradient-colored from blue-to-white-to-magenta.
Comment in
- Nup120: one more piece in the NPC puzzle.
Vetter IR. Vetter IR. Structure. 2009 Aug 12;17(8):1036-8. doi: 10.1016/j.str.2009.07.005. Structure. 2009. PMID: 19679081
Similar articles
- Molecular architecture of the Nup84-Nup145C-Sec13 edge element in the nuclear pore complex lattice.
Brohawn SG, Schwartz TU. Brohawn SG, et al. Nat Struct Mol Biol. 2009 Nov;16(11):1173-7. doi: 10.1038/nsmb.1713. Epub 2009 Oct 25. Nat Struct Mol Biol. 2009. PMID: 19855394 Free PMC article. - Structural and functional analysis of Nup120 suggests ring formation of the Nup84 complex.
Seo HS, Ma Y, Debler EW, Wacker D, Kutik S, Blobel G, Hoelz A. Seo HS, et al. Proc Natl Acad Sci U S A. 2009 Aug 25;106(34):14281-6. doi: 10.1073/pnas.0907453106. Epub 2009 Aug 11. Proc Natl Acad Sci U S A. 2009. PMID: 19706512 Free PMC article. - Nup120: one more piece in the NPC puzzle.
Vetter IR. Vetter IR. Structure. 2009 Aug 12;17(8):1036-8. doi: 10.1016/j.str.2009.07.005. Structure. 2009. PMID: 19679081 - Toward the atomic structure of the nuclear pore complex: when top down meets bottom up.
Hoelz A, Glavy JS, Beck M. Hoelz A, et al. Nat Struct Mol Biol. 2016 Jul;23(7):624-30. doi: 10.1038/nsmb.3244. Epub 2016 Jun 6. Nat Struct Mol Biol. 2016. PMID: 27273515 Free PMC article. Review. - Membrane-coating lattice scaffolds in the nuclear pore and vesicle coats: commonalities, differences, challenges.
Leksa NC, Schwartz TU. Leksa NC, et al. Nucleus. 2010 Jul-Aug;1(4):314-8. doi: 10.4161/nucl.1.4.11798. Epub 2010 Mar 12. Nucleus. 2010. PMID: 21327078 Free PMC article. Review.
Cited by
- Structural evolution of the membrane-coating module of the nuclear pore complex.
Liu X, Mitchell JM, Wozniak RW, Blobel G, Fan J. Liu X, et al. Proc Natl Acad Sci U S A. 2012 Oct 9;109(41):16498-503. doi: 10.1073/pnas.1214557109. Epub 2012 Sep 26. Proc Natl Acad Sci U S A. 2012. PMID: 23019579 Free PMC article. - The yeast nuclear pore complex and transport through it.
Aitchison JD, Rout MP. Aitchison JD, et al. Genetics. 2012 Mar;190(3):855-83. doi: 10.1534/genetics.111.127803. Genetics. 2012. PMID: 22419078 Free PMC article. - Scaffold nucleoporins Nup188 and Nup192 share structural and functional properties with nuclear transport receptors.
Andersen KR, Onischenko E, Tang JH, Kumar P, Chen JZ, Ulrich A, Liphardt JT, Weis K, Schwartz TU. Andersen KR, et al. Elife. 2013 Jun 11;2:e00745. doi: 10.7554/eLife.00745. Elife. 2013. PMID: 23795296 Free PMC article. - Molecular architecture of the Nup84-Nup145C-Sec13 edge element in the nuclear pore complex lattice.
Brohawn SG, Schwartz TU. Brohawn SG, et al. Nat Struct Mol Biol. 2009 Nov;16(11):1173-7. doi: 10.1038/nsmb.1713. Epub 2009 Oct 25. Nat Struct Mol Biol. 2009. PMID: 19855394 Free PMC article. - The nuclear pore complex has entered the atomic age.
Brohawn SG, Partridge JR, Whittle JR, Schwartz TU. Brohawn SG, et al. Structure. 2009 Sep 9;17(9):1156-68. doi: 10.1016/j.str.2009.07.014. Structure. 2009. PMID: 19748337 Free PMC article. Review.
References
- Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58:1948–1954. - PubMed
- Alber F, Dokudovskaya S, Veenhoff LM, Zhang W, Kipper J, Devos D, Suprapto A, Karni-Schmidt O, Williams R, Chait BT, et al. The molecular architecture of the nuclear pore complex. Nature. 2007;450:695–701. - PubMed
- Beck M, Lucic V, Forster F, Baumeister W, Medalia O. Snapshots of nuclear pore complexes in action captured by cryo-electron tomography. Nature. 2007;449:611–615. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases