Nuclear pore proteins and cancer - PubMed (original) (raw)
Review
Nuclear pore proteins and cancer
Songli Xu et al. Semin Cell Dev Biol. 2009 Jul.
Abstract
Nucleocytoplasmic trafficking of macromolecules, a highly specific and tightly regulated process, occurs exclusively through the nuclear pore complex. This immense structure is assembled from approximately 30 proteins, termed nucleoporins. Here we discuss the four nucleoporins that have been linked to cancers, either through elevated expression in tumors (Nup88) or through involvement in chromosomal translocations that encode chimeric fusion proteins (Tpr, Nup98, Nup214). In each case we consider the normal function of the nucleoporin and its translocation partners, as well as what is known about their mechanistic contributions to carcinogenesis, particularly in leukemias. Studies of nucleoporin-linked cancers have revealed novel mechanisms of oncogenesis and in the future, should continue to expand our understanding of cancer biology.
Figures
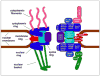
Figure 1. Structural organization of the Nuclear Pore Complex
Left: a cutaway representation of the organization of the NPC with the major structural elements indicated. Three of the eight repeating segments that give the NPC 8-fold rotational symmetry are illustrated. Right: Schematic of nucleoporin subcomplexes and approximate position within the NPC. Substructures are color coded to match the left side of the panel.
Figure 2. Nucleoporin structure and rearrangement following chromosomal translocations
The four nucleoporins discussed are depicted with major structural elements, along with their translocation partners, if applicable, and the structure of the resulting chimeric fusion proteins. Abbreviations: β-prop, β-propeller; c-c, coiled-coil; RTK, receptor tyrosine kinase; P/S, proline, serine rich; TM, transmembrane; JM, juxtamembrane; HD, homeodomain; BD, binding domain; Ub, ubiquitination; P, phosphorylation.
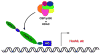
Figure 3. Model for Nup98 fusions
Nup98 fusion proteins are proposed to interact with chromatin directly through their homeodomain as depicted here or indirectly through the binding activity of otherfusion partners, e.g. NSD1 as part of a histone methyltransferase complex. The FG/GLFG repeat domain recruits chromatin modifying complexes such as CBP/p300 or HDAC1 which leads to altered transcription of target genes. The basis for target gene selection is not understood.
Similar articles
- Nucleoporins and nucleocytoplasmic transport in hematologic malignancies.
Takeda A, Yaseen NR. Takeda A, et al. Semin Cancer Biol. 2014 Aug;27:3-10. doi: 10.1016/j.semcancer.2014.02.009. Epub 2014 Mar 18. Semin Cancer Biol. 2014. PMID: 24657637 Review. - Characterization of the role of the tumor marker Nup88 in mitosis.
Hashizume C, Nakano H, Yoshida K, Wong RW. Hashizume C, et al. Mol Cancer. 2010 May 24;9:119. doi: 10.1186/1476-4598-9-119. Mol Cancer. 2010. PMID: 20497554 Free PMC article. - Moonlighting nuclear pore proteins: tissue-specific nucleoporin function in health and disease.
Jühlen R, Fahrenkrog B. Jühlen R, et al. Histochem Cell Biol. 2018 Dec;150(6):593-605. doi: 10.1007/s00418-018-1748-8. Epub 2018 Oct 25. Histochem Cell Biol. 2018. PMID: 30361777 Review. - Disruption of the FG nucleoporin NUP98 causes selective changes in nuclear pore complex stoichiometry and function.
Wu X, Kasper LH, Mantcheva RT, Mantchev GT, Springett MJ, van Deursen JM. Wu X, et al. Proc Natl Acad Sci U S A. 2001 Mar 13;98(6):3191-6. doi: 10.1073/pnas.051631598. Proc Natl Acad Sci U S A. 2001. PMID: 11248054 Free PMC article. - Nup98 localizes to both nuclear and cytoplasmic sides of the nuclear pore and binds to two distinct nucleoporin subcomplexes.
Griffis ER, Xu S, Powers MA. Griffis ER, et al. Mol Biol Cell. 2003 Feb;14(2):600-10. doi: 10.1091/mbc.e02-09-0582. Mol Biol Cell. 2003. PMID: 12589057 Free PMC article.
Cited by
- Rhinovirus 3C protease facilitates specific nucleoporin cleavage and mislocalisation of nuclear proteins in infected host cells.
Walker EJ, Younessi P, Fulcher AJ, McCuaig R, Thomas BJ, Bardin PG, Jans DA, Ghildyal R. Walker EJ, et al. PLoS One. 2013 Aug 7;8(8):e71316. doi: 10.1371/journal.pone.0071316. eCollection 2013. PLoS One. 2013. PMID: 23951130 Free PMC article. - Nup98 regulation of histone methylation promotes normal gene expression and may drive leukemogenesis.
Sump B, Brickner JH. Sump B, et al. Genes Dev. 2017 Nov 15;31(22):2201-2203. doi: 10.1101/gad.310359.117. Genes Dev. 2017. PMID: 29284709 Free PMC article. Review. - Molecular basis for the anchoring of proto-oncoprotein Nup98 to the cytoplasmic face of the nuclear pore complex.
Stuwe T, von Borzyskowski LS, Davenport AM, Hoelz A. Stuwe T, et al. J Mol Biol. 2012 Jun 22;419(5):330-46. doi: 10.1016/j.jmb.2012.03.024. Epub 2012 Apr 2. J Mol Biol. 2012. PMID: 22480613 Free PMC article. - Nucleoporin Nup188 is required for chromosome alignment in mitosis.
Itoh G, Sugino S, Ikeda M, Mizuguchi M, Kanno S, Amin MA, Iemura K, Yasui A, Hirota T, Tanaka K. Itoh G, et al. Cancer Sci. 2013 Jul;104(7):871-9. doi: 10.1111/cas.12159. Epub 2013 May 2. Cancer Sci. 2013. PMID: 23551833 Free PMC article. - Nuclear envelope budding and its cellular functions.
Keuenhof KS, Kohler V, Broeskamp F, Panagaki D, Speese SD, Büttner S, Höög JL. Keuenhof KS, et al. Nucleus. 2023 Dec;14(1):2178184. doi: 10.1080/19491034.2023.2178184. Nucleus. 2023. PMID: 36814098 Free PMC article.
References
- Tran EJ, Wente SR. Dynamic nuclear pore complexes: life on the edge. Cell. 2006;125(6):1041–53. - PubMed
- Schwartz TU. Modularity within the architecture of the nuclear pore complex. Curr Opin Struct Biol. 2005;15(2):221–6. - PubMed
- Terry LJ, Shows EB, Wente SR. Crossing the nuclear envelope: hierarchical regulation of nucleocytoplasmic transport. Science. 2007;318(5855):1412–6. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources