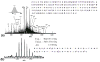Top-down proteomics reveals novel protein forms expressed in Methanosarcina acetivorans - PubMed (original) (raw)
Top-down proteomics reveals novel protein forms expressed in Methanosarcina acetivorans
Jonathan T Ferguson et al. J Am Soc Mass Spectrom. 2009 Sep.
Abstract
Using both automated nanospray and online liquid chromatography mass spectrometry LC-MS strategies, 99 proteins have been newly identified by top-down tandem mass spectrometry (MS/MS) in Methanosarcina acetivorans, the methanogen with the largest known genome [5.7 mega base pairs (Mb)] for an Archaeon. Because top-down MS/MS was used, 15 proteins were detected with mispredicted start sites along with an additional five from small open reading frames (SORFs). Beyond characterization of these more common discrepancies in genome annotation, one SORF resulted from a rare start codon (AUA) as the initiation site for translation of this protein. Also, a methylation on a 30S ribosomal protein (MA1259) was localized to Pro59-Val69, contrasting sharply from its homologue in Escherichia coli (rp S12) known to harbor an unusual beta-thiomethylated aspartic acid residue.
Figures
Figure 1
Mercury ion binding protein exhibiting a C-X-X-C motif was detected eluting at ~25 min (a). LC-MS/MS identified the protein (b) with an e-value of 4 × 10−11 (c). A disulfide bond was observed producing a −2 Da mass discrepancy. Thioredoxin has a C-X-X-C motif likely for facilitating reduction. Furthermore, this protein has a mispredicted start site (d), where the first four amino acids have been incorrectly annotated. The most abundant fragment ions have been labeled in (b).
Figure 2
Several proteins were detected whose genes are present on the chromosome in a putative operon in an LC-MS run (a). Using CID to produce b- and y-ions, each protein was identified with a mispredicted start site, (b)–(e).
Figure 3
A protein was identified despite not being annotated in the M. acetivorans database. In (a), 25 CID fragmentation scans of the LC-MS target were acquired via automated nanospray. A biomarker search of a database created from simple six-frame translations revealed a single hit, (b). DNA sequencing revealed that a Met (AUA) was the start codon for translation initiation. Four more unannotated proteins were identified using a biomarker search of a six-frame translated database, (c)–(f).
Figure 4
Intact protein MA1259 ribosomal protein S12P exhibits a +13.98 Da mass shift. Presumably, the uncalibrated data from the Q-FT mass spectrometer, which lacks automatic gain control, produced a −0.04 Da (−7.92 ppm) discrepancy from the theoretical +14.02 Da methylation. The protein was fragmented with ECD to localize the modification to a stretch of 30 amino acids (a). Offline analysis of a Glu-C digest detected a peptide with two missed cleavages incorporating the +14.02 Da modification (b). Fragmentation data further narrowed down possible sites of modification to 28 amino acids.
Similar articles
- Combination of Bottom-up 2D-LC-MS and Semi-top-down GelFree-LC-MS Enhances Coverage of Proteome and Low Molecular Weight Short Open Reading Frame Encoded Peptides of the Archaeon Methanosarcina mazei.
Cassidy L, Prasse D, Linke D, Schmitz RA, Tholey A. Cassidy L, et al. J Proteome Res. 2016 Oct 7;15(10):3773-3783. doi: 10.1021/acs.jproteome.6b00569. Epub 2016 Sep 2. J Proteome Res. 2016. PMID: 27557128 - Experimental determination of translational starts using peptide mass mapping and tandem mass spectrometry within the proteome of Mycobacterium tuberculosis.
Rison SCG, Mattow J, Jungblut PR, Stoker NG. Rison SCG, et al. Microbiology (Reading). 2007 Feb;153(Pt 2):521-528. doi: 10.1099/mic.0.2006/001537-0. Microbiology (Reading). 2007. PMID: 17259624 Free PMC article. - Multidimensional separation schemes enhance the identification and molecular characterization of low molecular weight proteomes and short open reading frame-encoded peptides in top-down proteomics.
Cassidy L, Helbig AO, Kaulich PT, Weidenbach K, Schmitz RA, Tholey A. Cassidy L, et al. J Proteomics. 2021 Jan 6;230:103988. doi: 10.1016/j.jprot.2020.103988. Epub 2020 Sep 17. J Proteomics. 2021. PMID: 32949814 - Quantitative proteome mapping of nitrotyrosines.
Bigelow DJ, Qian WJ. Bigelow DJ, et al. Methods Enzymol. 2008;440:191-205. doi: 10.1016/S0076-6879(07)00811-7. Methods Enzymol. 2008. PMID: 18423218 Review. - 2DE: the phoenix of proteomics.
Oliveira BM, Coorssen JR, Martins-de-Souza D. Oliveira BM, et al. J Proteomics. 2014 Jun 2;104:140-50. doi: 10.1016/j.jprot.2014.03.035. Epub 2014 Apr 3. J Proteomics. 2014. PMID: 24704856 Review.
Cited by
- Proteoforms expand the world of microproteins and short open reading frame-encoded peptides.
Cassidy L, Kaulich PT, Tholey A. Cassidy L, et al. iScience. 2023 Jan 27;26(2):106069. doi: 10.1016/j.isci.2023.106069. eCollection 2023 Feb 17. iScience. 2023. PMID: 36818287 Free PMC article. Review. - Extreme challenges and advances in archaeal proteomics.
Maupin-Furlow JA, Humbard MA, Kirkland PA. Maupin-Furlow JA, et al. Curr Opin Microbiol. 2012 Jun;15(3):351-6. doi: 10.1016/j.mib.2012.02.002. Epub 2012 Mar 1. Curr Opin Microbiol. 2012. PMID: 22386447 Free PMC article. Review. - Identification of a chloroform-soluble membrane miniprotein in Escherichia coli and its homolog in Salmonella typhimurium.
Guan Z, Wang X, Raetz CR. Guan Z, et al. Anal Biochem. 2011 Feb 15;409(2):284-9. doi: 10.1016/j.ab.2010.10.035. Epub 2010 Nov 2. Anal Biochem. 2011. PMID: 21050835 Free PMC article. - Molecular characterization of the thioredoxin system from Methanosarcina acetivorans.
McCarver AC, Lessner DJ. McCarver AC, et al. FEBS J. 2014 Oct;281(20):4598-611. doi: 10.1111/febs.12964. Epub 2014 Sep 6. FEBS J. 2014. PMID: 25112424 Free PMC article. - Sub-part-per-million precursor and product mass accuracy for high-throughput proteomics on an electron transfer dissociation-enabled orbitrap mass spectrometer.
Wenger CD, McAlister GC, Xia Q, Coon JJ. Wenger CD, et al. Mol Cell Proteomics. 2010 May;9(5):754-63. doi: 10.1074/mcp.M900541-MCP200. Epub 2010 Feb 2. Mol Cell Proteomics. 2010. PMID: 20124352 Free PMC article.
References
- Eichler J. Facing extremes: Archaeal surface-layer (glyco)proteins. Microbiology. 2003;149:3347–3351. - PubMed
- Daniels L. Biotechnological potential of methanogens. Biochem Soc Symp. 1992;58:181–193. - PubMed
- Galagan JE, Nusbaum C, Roy A, Endrizzi MG, Macdonald P, FitzHugh W, Calvo S, Engels R, Smirnov S, Atnoor D, Brown A, Allen N, Naylor J, Stange-Thomann N, DeArellano K, Johnson R, Linton L, McEwan P, McKernan K, Talamas J, Tirrell A, Ye W, Zimmer A, Barber RD, Cann I, Graham DE, Grahame DA, Guss AM, Hedderich R, Ingram-Smith C, Kuettner HC, Krzycki JA, Leigh JA, Li W, Liu J, Mukhopadhyay B, Reeve JN, Smith K, Springer TA, Umayam LA, White O, White RH, Conway de Macario E, Ferry JG, Jarrell KF, Jing H, Macario AJ, Paulsen I, Pritchett M, Sowers KR, Swanson RV, Zinder SH, Lander E, Metcalf WW, Birren B. The genome of M. acetivorans reveals extensive metabolic and physiological diversity. Genome Res. 2002;12:532–542. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous