Spp382p interacts with multiple yeast splicing factors, including possible regulators of Prp43 DExD/H-Box protein function - PubMed (original) (raw)
Spp382p interacts with multiple yeast splicing factors, including possible regulators of Prp43 DExD/H-Box protein function
Shatakshi Pandit et al. Genetics. 2009 Sep.
Abstract
Prp43p catalyzes essential steps in pre-mRNA splicing and rRNA biogenesis. In splicing, Spp382p stimulates the Prp43p helicase to dissociate the postcatalytic spliceosome and, in some way, to maintain the integrity of the spliceosome assembly. Here we present a dosage interference assay to identify Spp382p-interacting factors by screening for genes that when overexpressed specifically inhibit the growth of a conditional lethal prp38-1 spliceosome assembly mutant in the spp382-1 suppressor background. Identified, among others, are genes encoding the established splicing factors Prp8p, Prp9p, Prp11p, Prp39p, and Yhc1p and two poorly characterized proteins with possible links to splicing, Sqs1p and Cwc23p. Sqs1p copurifies with Prp43p and is shown to bind Prp43p and Spp382p in the two-hybrid assay. Overexpression of Sqs1p blocks pre-mRNA splicing and inhibits Prp43p-dependent steps in rRNA processing. Increased Prp43p levels buffer Sqs1p cytotoxicity, providing strong evidence that the Prp43p DExD/H-box protein is a target of Sqs1p. Cwc23p is the only known yeast splicing factor with a DnaJ motif characteristic of Hsp40-like chaperones. We show that similar to SPP382, CWC23 activity is critical for efficient pre-mRNA splicing and intron metabolism yet, surprisingly, this activity does not require the canonical DnaJ/Hsp40 motif. These and related data establish the value of this dosage interference assay for finding genes that alter cellular splicing and define Sqs1p and Cwc23p as prospective modulators of Spp382p-stimuated Prp43p function.
Figures
Figure 1.—
Schematic of the dosage interference assay. This diagram illustrates two possible fates of a spliceosome, completion of the splicing reaction and dissociation through Spp382p-stimulated Prp43p disassembly. Prp38-1p temperature inactivation is envisioned to favor complex dissociation (Kdis) over splicing (Ksp). The spp382-1 suppressor allele is shown to restore balance toward splicing in the prp38-1 mutant. This mutation also reduces the effectiveness of Prp43p in spliceosome disassembly, resulting in the accumulation of excess excised intron RNA. We screened a collection of yeast genes expressed by the robust GAL1 promoter to identify factors (large stars) that inhibit growth through interference with Spp382-1p-based suppression of prp38-1 or the natural Spp382p function in splicing.
Figure 2.—
Impact of gene overexpression on yeast growth. (A) The spp382-1 suppressed prp38-1 spliceosome assembly mutant strain (prp38-1 spp382-1), wild-type yeast (PRP38 SPP382), and the spp382-1 and prp38-1 single-mutant stains were transformed with the indicated _GAL1_-driven constructs and assayed for growth on rich nutrient medium in galactose (YP Gal) or glucose (YP Glu) at the indicated temperatures. The unsuppressed prp38-1 allele supports growth at 32° but not at 35°. Spp382-1 is growth impaired at 35°. To facilitate comparison between culture conditions, the plates were incubated for 2–6 days until the fastest growing strain in each set reached a colony size similar to the vector-only control transformant (Vector) in the prp38-1 spp382-1 background. (B) A growth assay of transformants in the wild-type PRP38 SPP382 (WT) background on defined synthetic medium containing galactose (−Ura Gal) or glucose (−Ura Glu) at 23° and 35°.
Figure 3.—
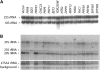
Splicing impairment with GAL1 gene expression. (A) RNA isolated from the indicated prp38-1 spp382-1 transformants after growth on galactose medium for 8 hr (the initial 6 hr at room temperature and the final 2 hr at 37°). The top panel shows hybridization for the intron-bearing RPS17A pre-RNA (P) and processed mRNA (M). The mRNA/pre-mRNA indicator of splicing efficiency is in parentheses (the values for SQS1 were too low to measure and are reported as not done, ND). Normalization for loading and general RNA destabilization is provided by hybridization for the intronless U2 snRNA. (B) RNA was isolated as in A except that yeast were grown continuously at 23°. The mRNA/pre-mRNA ratio is noted above the lanes. (C) RNA from yeast that express GAL1-PRP43 was resolved before (no oligo) and after treatment with RNase H and an oligonucleotide complementary to the poly(A) tail [oligo(dT)] or a sequence not present in the RPS17A transcript (control oligo). The unprocessed RPS17A precursor dimer (Pre-mRNA, brackets) and mRNA are shown at the left.
Figure 4.—
GAL1-SQS1 expression inhibits rRNA processing. Hybridization of the membrane transfer described in Figure 3A is shown with oligonucleotides specific for the 35S rRNA precursor and the 27SA2, 23S, and 20S rRNA processing intermediates. An unidentified background band seen with the 27SA2 probe provides an additional normalization control.
Figure 5.—
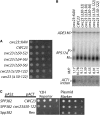
GAL1-SQS1 inhibits pre-mRNA splicing in wild-type yeast and interacts with Prp43p. (A) Northern analysis of RNA isolated from wild-type yeast transformed _GAL1_-driven copies of the five yeast G-patch protein genes (SPP2, SPP382, SQS1, PXR1, and YLR271W) and an empty vector. The cultures were grown on galactose medium for 8 hr (the initial 6 hr at room temperature and the final 2 hr at 37°; equivalent results were obtained when the cultures were grown continuously at 30°). The positions of the RPS17A precursor (P) and spliced mRNA (M) and the mRNA/pre-mRNA ratio are noted (top panel) and the 23S and 20S ribosomal RNA intermediates are shown (bottom panel). (B) Northern analysis of RNA isolated from wild-type yeast (WT), the sqs1∷KAN strain, and the splicing-impaired spp382-6 mutant (P
andit
et al. 2006) grown continuously at 23° (−) or after a 2-hr shift to 37° (+). The positions of RPS17A pre-mRNA, mRNA, and _ADE_3 mRNA are indicated at the left. The mRNA/pre-mRNA ratio is in parentheses. (C) Growth of yeast transformed with the indicated constructs on medium that induces (galactose) or represses (glucose) GAL1 expression for 3 days at 30° (except for the GAL1-SQS1 pPRP43 double transformant and its matched GAL1-SQS1 single transformant that incubated for 15 days). (D) Two-hybrid assays conducted on medium lacking histidine supplemented with 20 m
m
3-aminotriazole for reporter gene activation (Y2H Reporter) or simply lacking leucine and tryptophan to score for the two-hybrid plasmids (Plasmid Marker). pACT is the Gal4 activator construct, pAS2 is the Gal4 DNA binding construct, and empty refers to an empty vector control.
Figure 6.—
Cwc23p is required for efficient pre-mRNA splicing but its DnaJ motif is not critical. (A) Growth of wild-type yeast (CWC23), the cwc23 null mutant (cwc23∷KAN), and yeast bearing the indicated J-motif mutations on synthetic defined medium at 30°. (B) Northern blot of RNA isolated from the yeast described in A probed to detect RPS17A pre-mRNA (P) and RPS17A mRNA (m) and intronless ADE3 mRNA (top panel) or the excised intron from the ACT1 gene (bottom panel). (C) Two-hybrid interaction between a full-length construct of Spp382p and full-length Cwc23p or the deletion derivative removing much of the DnaJ domain (cwc23_Δ_50-122). Rev indicates a control construct in which the CWC23 open reading frame was inserted in the opposite orientation to the GAL4 sequence of the two-hybrid vector. The yeast strains were scored for reporter gene activation on medium lacking adenine.
Similar articles
- Limited portability of G-patch domains in regulators of the Prp43 RNA helicase required for pre-mRNA splicing and ribosomal RNA maturation in Saccharomyces cerevisiae.
Banerjee D, McDaniel PM, Rymond BC. Banerjee D, et al. Genetics. 2015 May;200(1):135-47. doi: 10.1534/genetics.115.176461. Epub 2015 Mar 25. Genetics. 2015. PMID: 25808954 Free PMC article. - Inhibition of a spliceosome turnover pathway suppresses splicing defects.
Pandit S, Lynn B, Rymond BC. Pandit S, et al. Proc Natl Acad Sci U S A. 2006 Sep 12;103(37):13700-5. doi: 10.1073/pnas.0603188103. Epub 2006 Aug 31. Proc Natl Acad Sci U S A. 2006. PMID: 16945917 Free PMC article. - The ATPase and helicase activities of Prp43p are stimulated by the G-patch protein Pfa1p during yeast ribosome biogenesis.
Lebaron S, Papin C, Capeyrou R, Chen YL, Froment C, Monsarrat B, Caizergues-Ferrer M, Grigoriev M, Henry Y. Lebaron S, et al. EMBO J. 2009 Dec 16;28(24):3808-19. doi: 10.1038/emboj.2009.335. EMBO J. 2009. PMID: 19927118 Free PMC article. - Helicases involved in splicing from malaria parasite Plasmodium falciparum.
Tuteja R. Tuteja R. Parasitol Int. 2011 Dec;60(4):335-40. doi: 10.1016/j.parint.2011.09.007. Epub 2011 Oct 1. Parasitol Int. 2011. PMID: 21996352 Review. - The current understanding of Ded1p/DDX3 homologs from yeast to human.
Tarn WY, Chang TH. Tarn WY, et al. RNA Biol. 2009 Jan-Mar;6(1):17-20. doi: 10.4161/rna.6.1.7440. Epub 2009 Jan 18. RNA Biol. 2009. PMID: 19106629 Review.
Cited by
- Limited portability of G-patch domains in regulators of the Prp43 RNA helicase required for pre-mRNA splicing and ribosomal RNA maturation in Saccharomyces cerevisiae.
Banerjee D, McDaniel PM, Rymond BC. Banerjee D, et al. Genetics. 2015 May;200(1):135-47. doi: 10.1534/genetics.115.176461. Epub 2015 Mar 25. Genetics. 2015. PMID: 25808954 Free PMC article. - Dissection of the factor requirements for spliceosome disassembly and the elucidation of its dissociation products using a purified splicing system.
Fourmann JB, Schmitzová J, Christian H, Urlaub H, Ficner R, Boon KL, Fabrizio P, Lührmann R. Fourmann JB, et al. Genes Dev. 2013 Feb 15;27(4):413-28. doi: 10.1101/gad.207779.112. Genes Dev. 2013. PMID: 23431055 Free PMC article. - Assembling a protein-protein interaction map of the SSU processome from existing datasets.
Lim YH, Charette JM, Baserga SJ. Lim YH, et al. PLoS One. 2011 Mar 10;6(3):e17701. doi: 10.1371/journal.pone.0017701. PLoS One. 2011. PMID: 21423703 Free PMC article. - DNAJC17 is localized in nuclear speckles and interacts with splicing machinery components.
Pascarella A, Ferrandino G, Credendino SC, Moccia C, D'Angelo F, Miranda B, D'Ambrosio C, Bielli P, Spadaro O, Ceccarelli M, Scaloni A, Sette C, De Felice M, De Vita G, Amendola E. Pascarella A, et al. Sci Rep. 2018 May 17;8(1):7794. doi: 10.1038/s41598-018-26093-1. Sci Rep. 2018. PMID: 29773831 Free PMC article. - Cwc23 is a component of the NTR complex and functions to stabilize Ntr1 and facilitate disassembly of spliceosome intermediates.
Su YL, Chen HC, Tsai RT, Lin PC, Cheng SC. Su YL, et al. Nucleic Acids Res. 2018 Apr 20;46(7):3764-3773. doi: 10.1093/nar/gky052. Nucleic Acids Res. 2018. PMID: 29390077 Free PMC article.
References
- Aravind, L., and E. V. Koonin, 1999. G-patch: a new conserved domain in eukaryotic RNA-processing proteins and type D retroviral polyproteins. Trends Biochem. Sci. 24: 342–344. - PubMed
- Bauerova-Zabranska, H., J. Stokrova, K. Strisovsky, E. Hunter, T. Ruml et al., 2005. The RNA binding G-patch domain in retroviral protease is important for infectivity and D-type morphogenesis of Mason-Pfizer monkey virus. J. Biol. Chem. 280: 42106–42112. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases