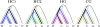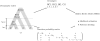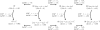Ancestral population genomics: the coalescent hidden Markov model approach - PubMed (original) (raw)
Ancestral population genomics: the coalescent hidden Markov model approach
Julien Y Dutheil et al. Genetics. 2009 Sep.
Abstract
With incomplete lineage sorting (ILS), the genealogy of closely related species differs along their genomes. The amount of ILS depends on population parameters such as the ancestral effective population sizes and the recombination rate, but also on the number of generations between speciation events. We use a hidden Markov model parameterized according to coalescent theory to infer the genealogy along a four-species genome alignment of closely related species and estimate population parameters. We analyze a basic, panmictic demographic model and study its properties using an extensive set of coalescent simulations. We assess the effect of the model assumptions and demonstrate that the Markov property provides a good approximation to the ancestral recombination graph. Using a too restricted set of possible genealogies, necessary to reduce the computational load, can bias parameter estimates. We propose a simple correction for this bias and suggest directions for future extensions of the model. We show that the patterns of ILS along a sequence alignment can be recovered efficiently together with the ancestral recombination rate. Finally, we introduce an extension of the basic model that allows for mutation rate heterogeneity and reanalyze human-chimpanzee-gorilla-orangutan alignments, using the new models. We expect that this framework will prove useful for population genomics and provide exciting insights into genome evolution.
Figures
Figure 1.—
The four different types that the genealogy of four sequences can take. The four species are H, human; C, chimpanzee; G, gorilla; and O, orangutan (outgroup). Following H
obolth
et al. (2007), these genealogies are labeled HC1, HC2, HG, and CG.
Figure 2.—
The genealogy of sequences varies along the genome. This graph shows the changes in topologies by colors (see Figure 1) together with the divergence between human and chimpanzee along 20,000 positions simulated using a coalescent with recombination (see
data and methods
).
Figure 3.—
The CoalHMM framework. The basic demographic model for human (H), chimpanzee (C), and gorilla (G) with the orangutan (O) as outgroup is used to design a set of candidate genealogies (topology plus coalescence times) and a corresponding transition matrix. These are computed according to the parameters of the model, namely speciation times (τ_X_), ancestral population sizes (θ_X_), and recombination rate ρ. The genealogies and transition probabilities are then used within a standard hidden Markov model to compute the likelihood and perform the posterior decoding.
Figure 4.—
Population and genealogy parameter notations.
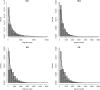
Figure 5.—
Population parameters recovery: (a) 07 implementation; (b–d) 09 implementation. (a and b) Ancestral recombination graph (ARG) simulated using a coalescent with recombination; (c) ARG simulated from the HMM; (d) ARG with averaged coalescent times. The dashed lines show the true values.
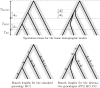
Figure 6.—
(a) Estimation of recombination rates. Ten data sets were simulated for each value of the recombination rate. The recombination rate was fixed to the same value in all lineages, and the 09 implementation with ρH = ρC = ρG was used for the estimation. Other parameters are set to the same values as in Figure 5. The dashed line shows the 1:1 ratio, and the solid line is fitted to the data. The slope is estimated to be 0.29. (b) Global vs. lineage-specific recombination rates. One hundred simulations were performed in each case, with parameter values identical to those in Figure 5. Dashed lines represent the true value of the recombination rate. Overlapping notches show nonsignificant differences in the medians.
Figure 7.—
Population parameters recovery when the bias correction is used. The parameter values used in the simulation are the same as in Figure 5.
Figure 8.—
Distribution of posterior fragment lengths. Shaded bars show the posterior distribution, hatched bars show the distribution from the ancestral recombination graph, and the solid lines show the geometric distribution (expected under the Markov assumption).
Figure 9.—
Comparison of the posterior decoding with the known ancestral recombination graph in 100 simulated data sets. (a) Joint distribution of reconstructed states (by column) and real states (by row). The numbers indicate the average percentage of each configuration. The proportions in all combinations sum to 100%, and the frequencies on the top-left/bottom-right diagonal correspond to the correctly inferred cases. The sums over each row give the true frequencies of each state, and the sums over columns provide the frequencies inferred by the CoalHMM method. (b) Precision and recall measures. The precision measure corresponds, for each column of panel 9a, to the diagonal proportion divided by the sum of the column (number of true positives over number of positives). The recall measure corresponds, for each row of panel 9a, to the diagonal proportion divided by the sum of the row (number of true positives over actual number of sites in a given genealogy). (c) Comparison of the reconstructed and real frequencies of hidden states. The proportion of incomplete lineage sorting is taken as the frequency of sites in alternative genealogies.
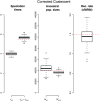
Figure 10.—
Model comparison on target 1 of the human–chimpanzee–gorilla–orangutan alignment. LRT: likelihood-ratio test. ΔBIC: difference in Bayesian information criterion. The arrows show the direction of the model comparison, pointing toward the alternative model. BIC is the difference in the BIC indexes between the two models. A negative value hence favors the alternative model, whereas a positive value favors the null model. NS: nonsignificant. ***P_-value <1_e − 16. Δ ln L: logarithm of the likelihood compared to the value of the 09 model. Γ: Gamma-distributed mutation rates.
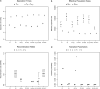
Figure 11.—
Parameter estimates obtained with different models applied to target 1. The parameter estimate for the gorilla recombination rate with the 09 model is 24 cM/Mb.
Similar articles
- Genomic relationships and speciation times of human, chimpanzee, and gorilla inferred from a coalescent hidden Markov model.
Hobolth A, Christensen OF, Mailund T, Schierup MH. Hobolth A, et al. PLoS Genet. 2007 Feb 23;3(2):e7. doi: 10.1371/journal.pgen.0030007. Epub 2006 Nov 30. PLoS Genet. 2007. PMID: 17319744 Free PMC article. - TRAILS: Tree reconstruction of ancestry using incomplete lineage sorting.
Rivas-González I, Schierup MH, Wakeley J, Hobolth A. Rivas-González I, et al. PLoS Genet. 2024 Feb 8;20(2):e1010836. doi: 10.1371/journal.pgen.1010836. eCollection 2024 Feb. PLoS Genet. 2024. PMID: 38330138 Free PMC article. - Estimation of hominoid ancestral population sizes under bayesian coalescent models incorporating mutation rate variation and sequencing errors.
Burgess R, Yang Z. Burgess R, et al. Mol Biol Evol. 2008 Sep;25(9):1979-94. doi: 10.1093/molbev/msn148. Epub 2008 Jul 4. Mol Biol Evol. 2008. PMID: 18603620 - Unraveling recombination rate evolution using ancestral recombination maps.
Munch K, Schierup MH, Mailund T. Munch K, et al. Bioessays. 2014 Sep;36(9):892-900. doi: 10.1002/bies.201400047. Epub 2014 Jul 14. Bioessays. 2014. PMID: 25043668 Free PMC article. Review. - Ancestral population genomics.
Dutheil JY, Hobolth A. Dutheil JY, et al. Methods Mol Biol. 2012;856:293-313. doi: 10.1007/978-1-61779-585-5_12. Methods Mol Biol. 2012. PMID: 22399464 Review.
Cited by
- Incomplete lineage sorting patterns among human, chimpanzee, and orangutan suggest recent orangutan speciation and widespread selection.
Hobolth A, Dutheil JY, Hawks J, Schierup MH, Mailund T. Hobolth A, et al. Genome Res. 2011 Mar;21(3):349-56. doi: 10.1101/gr.114751.110. Epub 2011 Jan 26. Genome Res. 2011. PMID: 21270173 Free PMC article. - Methods and models for unravelling human evolutionary history.
Schraiber JG, Akey JM. Schraiber JG, et al. Nat Rev Genet. 2015 Dec;16(12):727-40. doi: 10.1038/nrg4005. Epub 2015 Nov 10. Nat Rev Genet. 2015. PMID: 26553329 Review. - Linking great apes genome evolution across time scales using polymorphism-aware phylogenetic models.
De Maio N, Schlötterer C, Kosiol C. De Maio N, et al. Mol Biol Evol. 2013 Oct;30(10):2249-62. doi: 10.1093/molbev/mst131. Epub 2013 Aug 1. Mol Biol Evol. 2013. PMID: 23906727 Free PMC article. - Blockwise HMM computation for large-scale population genomic inference.
Paul JS, Song YS. Paul JS, et al. Bioinformatics. 2012 Aug 1;28(15):2008-15. doi: 10.1093/bioinformatics/bts314. Epub 2012 May 28. Bioinformatics. 2012. PMID: 22641715 Free PMC article. - Estimating divergence time and ancestral effective population size of Bornean and Sumatran orangutan subspecies using a coalescent hidden Markov model.
Mailund T, Dutheil JY, Hobolth A, Lunter G, Schierup MH. Mailund T, et al. PLoS Genet. 2011 Mar;7(3):e1001319. doi: 10.1371/journal.pgen.1001319. Epub 2011 Mar 3. PLoS Genet. 2011. PMID: 21408205 Free PMC article.
References
- Burgess, R., and Z. Yang, 2008. Estimation of hominoid ancestral population sizes under Bayesian coalescent models incorporating mutation rate variation and sequencing errors. Mol. Biol. Evol. 25: 1979–1994. - PubMed
- Durbin, R., S. Eddy, A. Krogh and G. Mitchison, 1998. Biological Sequence Analysis: Probabilistic Models of Proteins and Nucleic Acids. Cambridge University Press, Cambridge/London/New York.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources