High definition profiling of mammalian DNA methylation by array capture and single molecule bisulfite sequencing - PubMed (original) (raw)
. 2009 Sep;19(9):1593-605.
doi: 10.1101/gr.095190.109. Epub 2009 Jul 6.
Andrew D Smith, Jude Kendall, Zhenyu Xuan, Kandasamy Ravi, Michelle Rooks, Michael Q Zhang, Kenny Ye, Arindam Bhattacharjee, Leonardo Brizuela, W Richard McCombie, Michael Wigler, Gregory J Hannon, James B Hicks
Affiliations
- PMID: 19581485
- PMCID: PMC2752124
- DOI: 10.1101/gr.095190.109
High definition profiling of mammalian DNA methylation by array capture and single molecule bisulfite sequencing
Emily Hodges et al. Genome Res. 2009 Sep.
Abstract
DNA methylation stabilizes developmentally programmed gene expression states. Aberrant methylation is associated with disease progression and is a common feature of cancer genomes. Presently, few methods enable quantitative, large-scale, single-base resolution mapping of DNA methylation states in desired regions of a complex mammalian genome. Here, we present an approach that combines array-based hybrid selection and massively parallel bisulfite sequencing to profile DNA methylation in genomic regions spanning hundreds of thousands of bases. This single molecule strategy enables methylation variable positions to be quantitatively examined with high sampling precision. Using bisulfite capture, we assessed methylation patterns across 324 randomly selected CpG islands (CGI) representing more than 25,000 CpG sites. A single lane of Illumina sequencing permitted methylation states to be definitively called for >90% of target sties. The accuracy of the hybrid-selection approach was verified using conventional bisulfite capillary sequencing of cloned PCR products amplified from a subset of the selected regions. This confirmed that even partially methylated states could be successfully called. A comparison of human primary and cancer cells revealed multiple differentially methylated regions. More than 25% of islands showed complex methylation patterns either with partial methylation states defining the entire CGI or with contrasting methylation states appearing in specific regional blocks within the island. We observed that transitions in methylation state often correlate with genomic landmarks, including transcriptional start sites and intron-exon junctions. Methylation, along with specific histone marks, was enriched in exonic regions, suggesting that chromatin states can foreshadow the content of mature mRNAs.
Figures
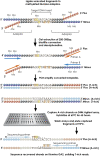
Figure 1.
Bisulfite capture procedure. Genomic DNA was randomly fragmented according to the standard Illumina protocol and ligated to custom-synthesized adaptors in which each C was replaced by 5-meC. The ligation was size-fractionated to select material from 150–300 bases in length. The gel-eluted material was treated with sodium bisulfite (see Methods) and then PCR-enriched using Illumina paired-end PCR primers. The resulting products were hybridized to custom-synthesized Agilent 244K arrays containing probes complementary to the A-rich strands. Hybridizations were carried out with Agilent array CGH buffers under standard conditions. After washing, captured fragments were eluted in water at 95°C and amplified again prior to quantification and sequencing on the Illumina GA2 platform.
Figure 2.
Mapping bisulfite treated reads. (A) Reads were mapped to the reference genome by minimizing the number of potential mismatches. Any T in a read incurred no penalty for aligning with a C in the genome, and any C in a read was penalized for aligning with a T in the genome. (B) Quality scores were converted to mismatch penalties by assigning a penalty of 0 to the consensus base, and penalizing non-consensus bases proportionately to the difference between their quality score and the consensus base score. A difference of 80 (representing the maximum possible range at a single position) was equated with a penalty of 1.
Figure 3.
Distribution of CpG methylation frequencies. (A) A pairwise comparison of methylation at individual CpG sites between the two samples is shown. (B,C) For each sample, scatter plots of the proportion methylated for each CpG (_x_-axis) and the subsequent neighboring CpG within an island (_y_-axis, CpG+1) is displayed. This analysis was restricted to those CpGs with at least 40 reads in both samples.
Figure 4.
Methylation status of bisulfite sequenced clones. (A–D) Four independent CGI loci are shown. Two histograms plot methylation frequencies at individual CpG sites for both the bisulfite capture data (upper) and the conventional bisulfite cloning data (lower) for all four loci. The block diagrams illustrate methylation state at each CpG site for each individually analyzed clone.
Figure 5.
Patterns of methylation in CpG islands. Graphical representation of methylation patterns in nine CpG islands. A pair of graphics represents each CpG island, one graphic for each sample (top, CHP-SKN-1; bottom, MDA-MB-231). Each graphic shows a pair of plots, both with bars indicating the amount of methylated (yellow) and unmethylated (blue) reads mapping over each CpG. The upper plot shows the absolute numbers of reads and spacing between CpGs. The lower plot shows the proportions of methylated and unmethylated reads. Confidence intervals are indicated in gray, and the yellow bar inside the confidence interval indicates the exact methylation frequency. Similar plots for the remaining CGIs are given in Supplemental Figure S5.
Figure 6.
Blocks of DNA methylation overlap exons, histone H3K36me3, and histone H3K4me2 marks. (A) An example of a CGI that overlaps multiple exons. Annotated gene tracks were downloaded from the UCSC Genome Browser. The gene tracks are displayed above a histogram plotting methylation frequencies at specific CpG sites positioned along the region shown. Absolute read counts and actual distance between CpG sites are depicted in the upper histogram, whereas the lower histogram shows the proportion of methylated and unmethylated Cs at each site. Boxes with dashed borders highlight blocks of methylation overlapping exons. The edges of the block are defined by the point at which the proportion of reads methylated is at least 0.5. (B) Two examples for which the distribution of histone marks along the CGI reflects DNA methylation status. To display the ChIP-seq data, a wiggle track was created for each histone mark by counting reads mapped in five-base windows across the genome.
Similar articles
- High density DNA methylation array with single CpG site resolution.
Bibikova M, Barnes B, Tsan C, Ho V, Klotzle B, Le JM, Delano D, Zhang L, Schroth GP, Gunderson KL, Fan JB, Shen R. Bibikova M, et al. Genomics. 2011 Oct;98(4):288-95. doi: 10.1016/j.ygeno.2011.07.007. Epub 2011 Aug 2. Genomics. 2011. PMID: 21839163 - Analyzing the cancer methylome through targeted bisulfite sequencing.
Lee EJ, Luo J, Wilson JM, Shi H. Lee EJ, et al. Cancer Lett. 2013 Nov 1;340(2):171-8. doi: 10.1016/j.canlet.2012.10.040. Epub 2012 Nov 28. Cancer Lett. 2013. PMID: 23200671 Free PMC article. Review. - Genome-wide DNA methylation profiling using Infinium® assay.
Bibikova M, Le J, Barnes B, Saedinia-Melnyk S, Zhou L, Shen R, Gunderson KL. Bibikova M, et al. Epigenomics. 2009 Oct;1(1):177-200. doi: 10.2217/epi.09.14. Epigenomics. 2009. PMID: 22122642 - Targeted bisulfite sequencing by solution hybrid selection and massively parallel sequencing.
Lee EJ, Pei L, Srivastava G, Joshi T, Kushwaha G, Choi JH, Robertson KD, Wang X, Colbourne JK, Zhang L, Schroth GP, Xu D, Zhang K, Shi H. Lee EJ, et al. Nucleic Acids Res. 2011 Oct;39(19):e127. doi: 10.1093/nar/gkr598. Epub 2011 Jul 23. Nucleic Acids Res. 2011. PMID: 21785137 Free PMC article. - Methods for CpG Methylation Array Profiling Via Bisulfite Conversion.
Leti F, Llaci L, Malenica I, DiStefano JK. Leti F, et al. Methods Mol Biol. 2018;1706:233-254. doi: 10.1007/978-1-4939-7471-9_13. Methods Mol Biol. 2018. PMID: 29423802 Free PMC article. Review.
Cited by
- Identifying Differential Methylation in Cancer Epigenetics via a Bayesian Functional Regression Model.
Shokoohi F, Stephens DA, Greenwood CMT. Shokoohi F, et al. Biomolecules. 2024 May 29;14(6):639. doi: 10.3390/biom14060639. Biomolecules. 2024. PMID: 38927043 Free PMC article. - scDMV: a zero-one inflated beta mixture model for DNA methylation variability with scBS-seq data.
Zhou Y, Zhang Y, Peng M, Zhang Y, Li C, Shu L, Hu Y, Su J, Xu J. Zhou Y, et al. Bioinformatics. 2024 Jan 2;40(1):btad772. doi: 10.1093/bioinformatics/btad772. Bioinformatics. 2024. PMID: 38141207 Free PMC article. - Current and Emerging Technologies for the Analysis of the Genome-Wide and Locus-Specific DNA Methylation Patterns.
Tost J. Tost J. Adv Exp Med Biol. 2022;1389:395-469. doi: 10.1007/978-3-031-11454-0_16. Adv Exp Med Biol. 2022. PMID: 36350519 - Enzymology of Mammalian DNA Methyltransferases.
Jurkowska RZ, Jeltsch A. Jurkowska RZ, et al. Adv Exp Med Biol. 2022;1389:69-110. doi: 10.1007/978-3-031-11454-0_4. Adv Exp Med Biol. 2022. PMID: 36350507 - Bacterial N4-methylcytosine as an epigenetic mark in eukaryotic DNA.
Rodriguez F, Yushenova IA, DiCorpo D, Arkhipova IR. Rodriguez F, et al. Nat Commun. 2022 Feb 28;13(1):1072. doi: 10.1038/s41467-022-28471-w. Nat Commun. 2022. PMID: 35228526 Free PMC article.
References
- Albert TJ, Molla MN, Muzny DM, Nazareth L, Wheeler D, Song X, Richmond TA, Middle CM, Rodesch MJ, Packard CJ, et al. Direct selection of human genomic loci by microarray hybridization. Nat Methods. 2007;4:903–905. - PubMed
- Bestor T, Laudano A, Mattaliano R, Ingram V. Cloning and sequencing of a cDNA encoding DNA methyltransferase of mouse cells. The carboxyl-terminal domain of the mammalian enzymes is related to bacterial restriction methyltransferases. J Mol Biol. 1988;203:971–983. - PubMed
- Bird AP. CpG-rich islands and the function of DNA methylation. Nature. 1986;321:209–213. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources