Histone methylation regulator PTIP is required for PPARgamma and C/EBPalpha expression and adipogenesis - PubMed (original) (raw)
Histone methylation regulator PTIP is required for PPARgamma and C/EBPalpha expression and adipogenesis
Young-Wook Cho et al. Cell Metab. 2009 Jul.
Abstract
PPARgamma and C/EBPalpha cooperate to control preadipocyte differentiation (adipogenesis). However, the factors that regulate PPARgamma and C/EBPalpha expression during adipogenesis remain largely unclear. Here, we show PTIP, a protein that associates with histone H3K4 methyltransferases, regulates PPARgamma and C/EBPalpha expression in mouse embryonic fibroblasts (MEFs) and during preadipocyte differentiation. PTIP deletion in MEFs leads to marked decreases of PPARgamma expression and PPARgamma-stimulated C/EBPalpha expression. Further, PTIP is essential for induction of PPARgamma and C/EBPalpha expression during preadipocyte differentiation. Deletion of PTIP impairs the enrichment of H3K4 trimethylation and RNA polymerase II on PPARgamma and C/EBPalpha promoters. Accordingly, PTIP(-/-) MEFs and preadipocytes all show striking defects in adipogenesis. Rescue of the adipogenesis defect in PTIP(-/-) MEFs requires coexpression of PPARgamma and C/EBPalpha. Finally, deletion of PTIP in brown adipose tissue significantly reduces tissue weight. Thus, by regulating PPARgamma and C/EBPalpha expression, PTIP plays a critical role in adipogenesis.
Figures
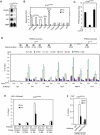
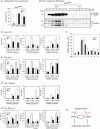
Figure 1. PTIP Is Required for PPARγ Expression in MEFs
(A) – (B) Immortalized _PTIP_flox/flox MEFs were infected with retrovirus MSCVpuro expressing Cre or vector (Vec) alone. Total RNA was extracted for confirmation of putative PTIP-regulated genes identified in microarray analysis (Supplemental Table S1). (A) Northern blot; (B) qRT-PCR. PPARγ2 mRNA level is below detection limit in qRT-PCR. AK5 and Flt1 stand for Adenylate Kinase 5 and FMS-like tyrosine kinase 1, respectively. 36B4 is the loading control. (C) Ectopic PTIP can rescue PPARγ expression in PTIP-deficient cells. _PTIP_flox/flox MEFs were sequentially infected with retroviruses MSCVhygro expressing Cre and MSCVpuro expressing FLAG-tagged PTIP (F-PTIP). PPARγ expression was analyzed by qRT-PCR. (D) ChIP assay of binding of the PTIP-associated HMT complex subunits to PPARγ1 and PPARγ2 promoters in _PTIP_flox/flox MEFs. Top panel, schematic of mouse PPARγ1 and PPARγ2 promoters. The black bars indicate locations of Taqman probes used for PCR quantitation of ChIP assays. Pol II, RNA polymerase II. (E) ChIP assay of H3K4me3 on PPARγ1 and PPARγ2 promoters in _PTIP_flox/flox MEFs. me3, H3K4me3. (F) ChIP assay of H3K4me3 and H3K27me3 on PPARγ1 promoter in _PTIP_flox/flox MEFs. All results are representative of two – four independent experiments.
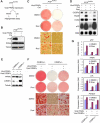
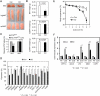
Figure 2. PTIP Is Required for Adipogenesis of Primary MEFs and Preadipocytes
(A) – (C) PTIP is required for adipogenesis of primary MEFs. Primary _PTIP_flox/flox MEFs were infected with adenovirus expressing Cre (Ad-Cre) or control virus expressing GFP alone, followed by adipogenesis assay. (A) To determine Ad-Cre-mediated gene deletion efficiency, PTIP expression was analyzed by qRT-PCR before induction of adipogenesis. (B) Morphological differentiation under the phase-contrast microscope (Oil Red O staining). (C) qRT-PCR analysis of gene expression after adipogenesis. (D) – (F) PTIP is required for differentiation of 3T3-L1 white preadipocyte cell line. 3T3-L1 cells were infected with pSUPER.retro-based retroviral shRNA construct targeting PTIP or vector alone. After puromycin selection, cells were induced to undergo adipogenesis for 8 days. (D) Western blot of PTIP expression before induction of differentiation. The asterisk indicates a non-specific band. (E) Morphological differentiation. (F) qRT-PCR analysis of gene expression 8 days after induction of differentiation. (G) – (L) PTIP is required for differentiation of primary white (G – I) and brown (J – L) preadipocytes. Primary white and brown preadipocytes isolated from _PTIP_flox/flox mice were infected with Ad-Cre or control virus expressing GFP, followed by adipogenesis assay. (G) and (J) qRT-PCR analysis of Cre-mediated PTIP gene deletion before differentiation. (H) and (K) Morphological differentiation. Top panels, stained dishes. Lower panels, representative fields under the microscope. (I) and (L) After differentiation, total RNA was isolated and subjected to qRT-PCR analysis of expression of genes common to white and brown adipocytes (PPARγ, PPARγ1, PPARγ2, C/EBPα, aP2) and genes specific for brown adipocytes (PRDM16, cidea, eva1, UCP1, PGC-1α) (Seale et al., 2007). All results are representative of two – four independent experiments.
Figure 3. Both PTIP-Deficient and C/EBPα-Deficient MEFs Show Defects in PPARγ-Stimulated Adipogenesis
(A) – (D) PTIP-deficient MEFs show significant defects in PPARγ-stimulated adipogenesis. Immortalized _PTIP_flox/flox MEFs expressing ectopic PPARγ were infected with MSCVpuro expressing Cre or vector alone, followed by adipogenesis assay in the presence or absence of synthetic PPARγ ligand Rosi. (A) Schematic of the PPARγ-stimulated adipogenesis assay. (B) Western blot of endogenous PTIP and ectopic PPARγ expression before adipogenesis. (C) Morphological differentiation (Oil Red O staining). Top panels, stained dishes. Lower panels, representative fields under the microscope. (D) Northern blot analysis of gene expression after adipogenesis. (E) – (G) C/EBPα-deficient MEFs show significant defects in PPARγ-stimulated adipogenesis. Immortalized C/EBPα+/− and _C/EBPα_−/− MEFs infected with MSCVpuro-PPARγ2 were then infected with MSCVhygro expressing C/EBPα, followed by adipogenesis assay for 8 days in the presence or absence of Rosi. (E) Western blot of PPARγ and C/EBPα expression before adipogenesis. (F) Morphological differentiation. (G) qRT-PCR analysis of gene expression after adipogenesis. PGAR is also known as Angiopoietin-like 4 (Angptl4). FASN stands for Fatty acid synthase. LPL stands for Lipoprotein lipase. All results are representative of two – four independent experiments.
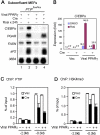
Figure 4. PTIP Directly Regulates C/EBPα Expression in MEFs
(A) – (B) PTIP-deficient MEFs show over 5 fold decrease of PPARγ- and ligand-stimulated expression of endogenous C/EBPα. MEFs described in Figure 3A-D were treated with 0.5μM Rosi or DMSO for 24 h, followed by Northern blot (A) and qRT-PCR analysis (B). When RNA samples were collected, cell density was less than 80−90%. (C) ChIP assay of PTIP recruitment to the C/EBPα promoter in _PTIP_flox/flox MEFs. (D) ChIP assay of H3K4me3 enrichment on the C/EBPα promoter in _PTIP_flox/flox MEFs. All results are representative of three independent experiments.
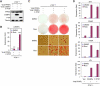
Figure 5. Rescue of the Adipogenesis Defect in _PTIP_−/− MEFs Requires Co-expression of PPARγ and C/EBPα
_PTIP_−/− MEFs were established by infecting immortalized _PTIP_flox/flox MEFs with Ad-Cre, followed by isolation of single colonies. _PTIP_−/− MEFs were infected with MSCVpuro expressing PPARγ, followed by infection with MSCVhygro expressing C/EBPα or F-PTIP. (A) Western blot analysis of ectopic PPARγ, C/EBPα and PTIP expression before induction of adipogenesis. Cells were cultured in the absence of Rosi. (B) Ectopic PTIP rescues PPARγ- and ligand-stimulated expression of endogenous C/EBPα in subconfluent, undifferentiated _PTIP_−/− MEFs. Cells were treated with 0.5 μM Rosi or DMSO for 24 h. C/EBPα expression was analyzed by qRT-PCR. (C) Cells were induced to undergo adipogenesis for 8 days and were stained with Oil Red O. (D) qRT-PCR analysis of gene expression after adipogenesis. All results are representative of two – four independent experiments.
Figure 6. PTIP Is Required for Induction of PPARγ and C/EBPα during Adipogenesis
(A) – (B) PTIP is essential for induction of PPARγ and C/EBPα expression during preadipocyte differentiation. Adipogenesis of primary _PTIP_flox/flox brown preadipocytes was performed as in Figure 2J-L. The basal level of PPARγ and C/EBPα expression in undifferentiated preadipocytes was determined by qRT-PCR (A). PPARγ and C/EBPα protein expression during preadipocyte differentiation was analyzed by Western blot (B). Whole cell extracts were collected every 24 h following induction of differentiation with IBMX, DEX and indomethacin at day 0. The 54.5 kDa PPARγ1 and 57.5 kDa PPARγ2 bands, as well as the 30 kDa and 42 kDa C/EBPα isoforms are indicated by arrows. The asterisk indicates a non-specific band. (C) qRT-PCR analysis of C/EBPβ expression during differentiation of _PTIP_flox/flox brown preadipocytes. RNA samples were collected every 24 h starting from three days before induction of adipogenesis (d-3). (D) – (G) ChIP assays of the recruitment of PTIP, MLL4 and Pol II, and H3K4me3 signal on PPARγ1, PPARγ2 and C/EBPα promoters in differentiating (day 4) _PTIP_flox/flox brown preadipocytes. (H) Model depicting the role of PTIP in transcriptional regulation of preadipocyte differentiation (adipogenesis). PTIP regulates adipogenesis through direct modulation of PPARγ and C/EBPα expression. All results are representative of two – four independent experiments.
Figure 7. Deletion of PTIP in Brown Adipose Tissue Reduces Tissue Weight and Leads to Cold Intolerance in Mice
(A) – (D) PTIP deletion in brown adipose tissue (BAT) decreases both tissue weight and expression of brown adipocyte-enriched genes. Adipose-specific PTIP knockout mice (KO) carried the genotype of _PTIP_flox/−;Cre. Other littermates were used as control (Con) (see Experimental Procedures). All data were from 12−16 week-old mice. (A) Representative pictures of interscapular BAT, inguinal WAT (ingWAT) and epididymal WAT (epiWAT) isolated from Con and KO mice. (B) Quantitation of BAT and WAT weights in Con (n = 22) and KO (n = 8) mice. No significant differences in body weight, BMI and adipose tissue weights were found among groups of control mice (Figure S4). (C) Lack of aP2-Cre-mediated deletion of PTIP gene in WAT of KO mice as determined by quantitative PCR (n = 8). (D) qRT-PCR analysis of gene expression in BAT of Con (n = 12) and KO (n = 8) mice. (E) – (F) PTIP deletion in BAT leads to cold intolerance in mice. (E) The body temperatures of mice housed in 4°C were measured every hour for 6h (n = 7−9). (F) qRT-PCR analysis of cold-induced gene expression in BAT of mice after cold treatment for 6h (n = 6). All values are the mean +/− SEM. *P < 0.05; **P < 0.01. Similar results were obtained from male and female mice but only the male mice data are shown here.
Similar articles
- MLL3/MLL4-Associated PAGR1 Regulates Adipogenesis by Controlling Induction of C/EBPβ and C/EBPδ.
Lee JE, Cho YW, Deng CX, Ge K. Lee JE, et al. Mol Cell Biol. 2020 Aug 14;40(17):e00209-20. doi: 10.1128/MCB.00209-20. Print 2020 Aug 14. Mol Cell Biol. 2020. PMID: 32601106 Free PMC article. - Cyclin C regulates adipogenesis by stimulating transcriptional activity of CCAAT/enhancer-binding protein α.
Song Z, Xiaoli AM, Zhang Q, Zhang Y, Yang EST, Wang S, Chang R, Zhang ZD, Yang G, Strich R, Pessin JE, Yang F. Song Z, et al. J Biol Chem. 2017 May 26;292(21):8918-8932. doi: 10.1074/jbc.M117.776229. Epub 2017 Mar 28. J Biol Chem. 2017. PMID: 28351837 Free PMC article. - Krüppel-like factor 10 (KLF10) is transactivated by the transcription factor C/EBPβ and involved in early 3T3-L1 preadipocyte differentiation.
Liu Y, Peng WQ, Guo YY, Liu Y, Tang QQ, Guo L. Liu Y, et al. J Biol Chem. 2018 Sep 7;293(36):14012-14021. doi: 10.1074/jbc.RA118.004401. Epub 2018 Jul 19. J Biol Chem. 2018. PMID: 30026232 Free PMC article. - Control of histone methylation and genome stability by PTIP.
Muñoz IM, Rouse J. Muñoz IM, et al. EMBO Rep. 2009 Mar;10(3):239-45. doi: 10.1038/embor.2009.21. Epub 2009 Feb 20. EMBO Rep. 2009. PMID: 19229280 Free PMC article. Review. - Role of histone methylation and demethylation in adipogenesis and obesity.
Okamura M, Inagaki T, Tanaka T, Sakai J. Okamura M, et al. Organogenesis. 2010 Jan-Mar;6(1):24-32. doi: 10.4161/org.6.1.11121. Organogenesis. 2010. PMID: 20592862 Free PMC article. Review.
Cited by
- Exploring the Genetic Conception of Obesity via the Dual Role of FoxO.
Behl T, Kaur I, Sehgal A, Singh S, Zengin G, Negrut N, Nistor-Cseppento DC, Pavel FM, Corb Aron RA, Bungau S. Behl T, et al. Int J Mol Sci. 2021 Mar 20;22(6):3179. doi: 10.3390/ijms22063179. Int J Mol Sci. 2021. PMID: 33804729 Free PMC article. Review. - Histone demethylase LSD1 regulates adipogenesis.
Musri MM, Carmona MC, Hanzu FA, Kaliman P, Gomis R, Párrizas M. Musri MM, et al. J Biol Chem. 2010 Sep 24;285(39):30034-41. doi: 10.1074/jbc.M110.151209. Epub 2010 Jul 23. J Biol Chem. 2010. PMID: 20656681 Free PMC article. - PTIP promotes chromatin changes critical for immunoglobulin class switch recombination.
Daniel JA, Santos MA, Wang Z, Zang C, Schwab KR, Jankovic M, Filsuf D, Chen HT, Gazumyan A, Yamane A, Cho YW, Sun HW, Ge K, Peng W, Nussenzweig MC, Casellas R, Dressler GR, Zhao K, Nussenzweig A. Daniel JA, et al. Science. 2010 Aug 20;329(5994):917-23. doi: 10.1126/science.1187942. Epub 2010 Jul 29. Science. 2010. PMID: 20671152 Free PMC article. - Review: Epigenetic regulation of adipocyte differentiation and adipogenesis.
Li HX, Xiao L, Wang C, Gao JL, Zhai YG. Li HX, et al. J Zhejiang Univ Sci B. 2010 Oct;11(10):784-91. doi: 10.1631/jzus.B0900401. J Zhejiang Univ Sci B. 2010. PMID: 20872986 Free PMC article. Review. - Novel insights into histone modifiers in adipogenesis.
Okuno Y, Inoue K, Imai Y. Okuno Y, et al. Adipocyte. 2013 Oct 1;2(4):285-8. doi: 10.4161/adip.25731. Epub 2013 Jul 16. Adipocyte. 2013. PMID: 24052908 Free PMC article.
References
- Abel ED, Peroni O, Kim JK, Kim Y-B, Boss O, Hadro E, Minnemann T, Shulman GI, Kahn BB. Adipose-selective targeting of the GLUT4 gene impairs insulin action in muscle and liver. Nature. 2001;409:729. - PubMed
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. A Bivalent Chromatin Structure Marks Key Developmental Genes in Embryonic Stem Cells. Cell. 2006;125:315. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- ZIADK047055-03/DK/NIDDK NIH HHS/United States
- ZIA DK075003-08/ImNIH/Intramural NIH HHS/United States
- ZIADK075017-01/DK/NIDDK NIH HHS/United States
- ZIADK075003-06/DK/NIDDK NIH HHS/United States
- Z01 DK047055-01/ImNIH/Intramural NIH HHS/United States
- Z01 DK075003-04/ImNIH/Intramural NIH HHS/United States
- Z01 DK047055-02/ImNIH/Intramural NIH HHS/United States
- Z99 DK999999/ImNIH/Intramural NIH HHS/United States
- Z01 DK075003-05/ImNIH/Intramural NIH HHS/United States
- ZIA DK075017-03/ImNIH/Intramural NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases