Proteomic analysis of murine Piwi proteins reveals a role for arginine methylation in specifying interaction with Tudor family members - PubMed (original) (raw)
Proteomic analysis of murine Piwi proteins reveals a role for arginine methylation in specifying interaction with Tudor family members
Vasily V Vagin et al. Genes Dev. 2009.
Abstract
In germ cells, Piwi proteins interact with a specific class of small noncoding RNAs, piwi-interacting RNAs (piRNAs). Together, these form a pathway that represses transposable elements, thus safeguarding germ cell genomes. Basic models describe the overall operation of piRNA pathways. However, the protein compositions of Piwi complexes, the critical protein-protein interactions that drive small RNA production and target recognition, and the precise molecular consequences of conserved localization to germline structures, call nuage, remains poorly understood. We purified the three murine Piwi family proteins, MILI, MIWI, and MIWI2, from mouse germ cells and characterized their interacting protein partners. Piwi proteins were found in complex with PRMT5/WDR77, an enzyme that dimethylates arginine residues. By immunoprecipitation with specific antibodies and by mass spectrometry, we found that Piwi proteins are arginine methylated at conserved positions in their N termini. These modifications are essential to direct complex formation with specific members of the Tudor protein family. Recognition of methylarginine marks by Tudor proteins can drive the localization of Piwi proteins to cytoplasmic foci in an artificial setting, supporting a role for this interaction in Piwi localization to nuage, a characteristic that correlates with proper operation of the piRNA pathway and transposon silencing in multiple organisms.
Figures
Figure 1.
PRMT5/WDR77 complex associate with mouse Piwi proteins. (A) Presented are MudPIT analyses of immuoprecipitated 3xFlag-HA-MIWI complexes with the number of peptides and coverage (percent shown in each column) shown for HA elution. (B, top panel) PRMT5 or WDR77 was immunoprecipitated from adult testis extracts and probed on a Western blot with HA antibodies to detect Flag-HA-MIWI. (Bottom panel) Flag-HA-tagged MIWI2 or MILI was immunoprecipitated from embryonic testes (E16.5–E18.5) using Flag beads and probed with PRMT5 or HA antibodies. (L) Lysate; (B) bound fractions. The bands seen in immunoprecipitates from nontransgenic animals represent a nonspecific interaction between PRMT5 and the anti-Flag beads. (C) Tagged MIWI, MIWI2, or MILI proteins were immunoprecipitated from 293 cell extracts using myc beads and probed on Westerns with HA antibodies.
Figure 2.
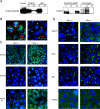
Arginine methylation of MIWI and the presence of arginine methylated proteins in the chromatoid body (CB). (A, left panel) Tagged MIWI protein was immunoprecipitaed from testes extracts with SYM10 (anti-symmetric dimethylarginine) and probed by Western with HA antibodies. (Right panel) Tagged MIWI was immunoprecipitated using Flag beads and tested on Western with SYM10 antibodies. (L) Lysate; (S) supernatant; (B) bound fractions. (B) Localization of symmetric dimethylarginine (SYM10) and MIWI was noted in the CB of round spermatids. Before the CB is formed, MIWI and Sym10 marks are not colocalized. In spermatocytes (Sp) MIWI is uniformly distributed in the cytoplasm, while Sym10 predominantly stain the nucleus. (C) Localization of symmetric (SYM10) and asymmetric (ASYM14) arginine methylation marks was analyzed in testes of 20-d-old heterozygous and homozygous Miwi animals. (D) The PRMT5 partner, WDR77 localizes to CB in control animals (Miwi+/−), but is lost in MIWI-deficient animals, while the CB markers, MVH and MILI, retain their localization in CBs. PRMT5 is uniformly distributed in the cytoplasm independently of MIWI.
Figure 3.
Mapping of arginine methylation sites in MIWI and MILI and conservation of arginine methylation sites among Piwi proteins. (A) Potential arginine methylation sites (RA/RG motifs) are shown at N termini of MILI and MIWI (RA motifs, green; RG motifs, red). Monomethylation (M) and dimethylation (D) sites were identified by three different methods using digestion with trypsin or V8 protease and color coded accordingly. Below are shown positions of four arginine residues changed to lysine in the MILI-m1 construct. (B) The N termini of murine (MIWI, MILI, and MIWI2), Drosophila (Piwi, Aub, and AGO3), cnidarian (Cniwi), and C. elegans (prg-1) Piwis and the Ago family members in Drosophila (dAgo1 and dAgo2) and mouse (mAgo2) are shown to scale. The positions of RA and RG motifs, marked as in A, are shown with stretches of multiple tandem motifs marked with the number of arginine residues. (Inset) Representative motifs in Piwis are aligned with known sites of symmetric arginine methylation in Sm D1, Sm D3, and p80 coilin. The boxed area of MILI and MIWI is shown in A.
Figure 4.
Piwi complexes with Tudor proteins. (A) Results of MudPIT analysis are shown for MILI complexes immunoprecipitated from adult testis extracts using MILI N1 or MILI N2 antibodies. The percent coverage is shown for each bar. (B) Results of MudPIT analysis are shown for MIWI complexes immunoprecipitated from adult testis using MIWI N2 or MIWI N3 antibodies. (C) Results of MudPIT analysis are shown for immuoprecipitated 3xFlag-HA-MIWI complexes (adult testis). (D, top panel) Results of MudPIT analysis of immunoprecipitated 3xFlag-HA-MIWI2 complexes from E16.5–E18.5 testes are shown. (Bottom panel) Results of MudPIT analysis of immunoprecipitated 3xFlag-HA-MILI complexes from RNaseA-treated extracts are shown. (E) 3xFlag-HA-MIWI was immunoprecipitaed from adult testis extracts using TDRD1, TDRD9, TDRD4, TDRD6, TDRD7, or IgG antibodies and probed on Western with HA antibodies. (F) 3xFlag-HA-MIWI2 or 3xFlag-HA-MILI were immunoprecipitated from embryonic testes (E16.5–E18.5) using Flag beads and tested on Western with TDRD1, TDRD9 antibodies. (L:) Lysate; (B) bound fractions.
Figure 5.
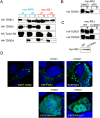
Tudors interacts with dimethylated arginines in Piwis. (A) Coimmunoprecipitation is shown for myc-MIWI or myc-MILI with Flag-HA-TDRD1, Flag-HA-TDRD2, Flag-HA-TDRD9, and Flag-HA-Tudor-SN from 293 cell extracts. Tagged MIWI or MILI proteins were immunoprecipitated from 293 cell with (+R) or without (−R) RNase A treatment using anti-myc beads and blotted using HA antibodies. (B) Coimmunoprecipitation is shown for myc-MILI with Flag-HA-TDRD1 from extracts of transfected 293 cells treated with an inhibitor of methylation (MTA). MILI was precipitated using anti-myc beads and probed with HA antibodies. (C, top panel) Coimmunoprecipitation is shown for Flag-HA-TDRD1 and TDRD9 with myc-MILI (WT) and myc-MILI-m1 (m1), where four arginine residues were substituted to lysine (see Fig. 3A). Tagged MILI proteins were immunoprecipitated from 293 cell extracts using anti-myc beads and probed with HA antibodies. (Bottom panel) The lysate, which was used for myc immunoprecipitation, was probed with myc antibodies. (D) Patterns of ectopically expressed TDRD1, MILI, and MILI-m1 localization are shown for U2OS cells. Also shown are patterns of MILI and MILI-m1 in U2OS cells that express a nontagged TDRD1.
Figure 6.
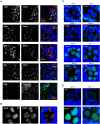
Colocalization and interdependence of Piwi and Tudor proteins in cytoplasmic nuage. (A) Localization of TDRD1, TDRD6, TDRD7, TDRD9 and MIWI are shown in testes of adult (2-mo-old) mice expressing the myc-MIWI transgene. Four Tudor proteins and MIWI colocalize in a single nuage granule, the chromatoid body, in round spermatids. TDRD1 and TDRD7 localize to numerous small nuage granules at earlier stages, primary spermatocytes (arrow), while MIWI is more dispersed in the cytoplasm at this stage. Note the nuclear localization of TDRD9 in spermatogonia and primary spermatocytes (arrowhead). (B) Localization of MIWI2 and TDRD9 in embryonic (E17.5) testes shows a complete colocalization of these proteins in a few cytoplasmic granules in prospermatogonia. (C) The effect of MILI and MIWI2 deficiency on TDRD1 and TDRD9 localization is shown for E17.5 prospermatogonia. (D) The effect of TDRD1 deficiency on MILI and MIWI2 localization is shown for E17.5 prospermatogonia.
Figure 7.
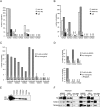
The effect of TDRD1 deficiency on piRNA expression in prospermatogonia. (A) Annotation is shown for small RNAs cloned from embryonic (E18.5) testes of Tdrd1 heterozygous and homozygous knockout (KO) animals. (B) Total cellular piRNA populations are composed of two complexes, MILI, with an average piRNA length 26 nt and MIWI2 with an average piRNA length 28 nt. The size profile reflects the ratio of both complexes in the cell. (C) The ratio of 28-nt (MIWI2) to 26-nt (MILI) piRNAs that correspond to LINE1 and IAP retrotransposons as well as single-stranded (cl1, chr.7) and double-stranded (cl2, chr.8) piRNA clusters decreases in the Tdrd1 mutant. (D) The fraction of secondary piRNAs matching retrotransposons and piRNA clusters decreases in Tdrd1 mutants. (E) The fraction of antisense piRNAs that correspond to L1 and IAP retrotransposons is reduced in Tdrd1 knockouts. (F) Localization of LINE1- and IAP-encoded proteins in testes of E17.5 heterozygous and homozygous Tdrd1 animals. Pictures of knockout and control animals were taken with identical exposure levels.
Similar articles
- Mouse Piwi interactome identifies binding mechanism of Tdrkh Tudor domain to arginine methylated Miwi.
Chen C, Jin J, James DA, Adams-Cioaba MA, Park JG, Guo Y, Tenaglia E, Xu C, Gish G, Min J, Pawson T. Chen C, et al. Proc Natl Acad Sci U S A. 2009 Dec 1;106(48):20336-41. doi: 10.1073/pnas.0911640106. Epub 2009 Nov 16. Proc Natl Acad Sci U S A. 2009. PMID: 19918066 Free PMC article. - PAPI, a novel TUDOR-domain protein, complexes with AGO3, ME31B and TRAL in the nuage to silence transposition.
Liu L, Qi H, Wang J, Lin H. Liu L, et al. Development. 2011 May;138(9):1863-73. doi: 10.1242/dev.059287. Epub 2011 Mar 29. Development. 2011. PMID: 21447556 Free PMC article. - Loss of the Mili-interacting Tudor domain-containing protein-1 activates transposons and alters the Mili-associated small RNA profile.
Reuter M, Chuma S, Tanaka T, Franz T, Stark A, Pillai RS. Reuter M, et al. Nat Struct Mol Biol. 2009 Jun;16(6):639-46. doi: 10.1038/nsmb.1615. Epub 2009 May 24. Nat Struct Mol Biol. 2009. PMID: 19465913 - How does the royal family of Tudor rule the PIWI-interacting RNA pathway?
Siomi MC, Mannen T, Siomi H. Siomi MC, et al. Genes Dev. 2010 Apr 1;24(7):636-46. doi: 10.1101/gad.1899210. Genes Dev. 2010. PMID: 20360382 Free PMC article. Review. - Deciphering arginine methylation: Tudor tells the tale.
Chen C, Nott TJ, Jin J, Pawson T. Chen C, et al. Nat Rev Mol Cell Biol. 2011 Sep 14;12(10):629-42. doi: 10.1038/nrm3185. Nat Rev Mol Cell Biol. 2011. PMID: 21915143 Review.
Cited by
- MIWI N-terminal RG motif promotes efficient pachytene piRNA production and spermatogenesis independent of LINE1 transposon silencing.
Wei C, Jing J, Yan X, Mann JM, Geng R, Xie H, Demireva EY, Hess RA, Ding D, Chen C. Wei C, et al. PLoS Genet. 2023 Nov 13;19(11):e1011031. doi: 10.1371/journal.pgen.1011031. eCollection 2023 Nov. PLoS Genet. 2023. PMID: 37956204 Free PMC article. - TDRD5 is required for retrotransposon silencing, chromatoid body assembly, and spermiogenesis in mice.
Yabuta Y, Ohta H, Abe T, Kurimoto K, Chuma S, Saitou M. Yabuta Y, et al. J Cell Biol. 2011 Mar 7;192(5):781-95. doi: 10.1083/jcb.201009043. J Cell Biol. 2011. PMID: 21383078 Free PMC article. - Minireview: The roles of small RNA pathways in reproductive medicine.
Hawkins SM, Buchold GM, Matzuk MM. Hawkins SM, et al. Mol Endocrinol. 2011 Aug;25(8):1257-79. doi: 10.1210/me.2011-0099. Epub 2011 May 5. Mol Endocrinol. 2011. PMID: 21546411 Free PMC article. Review. - HSP90α plays an important role in piRNA biogenesis and retrotransposon repression in mouse.
Ichiyanagi T, Ichiyanagi K, Ogawa A, Kuramochi-Miyagawa S, Nakano T, Chuma S, Sasaki H, Udono H. Ichiyanagi T, et al. Nucleic Acids Res. 2014 Oct 29;42(19):11903-11. doi: 10.1093/nar/gku881. Epub 2014 Sep 27. Nucleic Acids Res. 2014. PMID: 25262350 Free PMC article. - Roles of MIWI, MILI and PLD6 in small RNA regulation in mouse growing oocytes.
Kabayama Y, Toh H, Katanaya A, Sakurai T, Chuma S, Kuramochi-Miyagawa S, Saga Y, Nakano T, Sasaki H. Kabayama Y, et al. Nucleic Acids Res. 2017 May 19;45(9):5387-5398. doi: 10.1093/nar/gkx027. Nucleic Acids Res. 2017. PMID: 28115634 Free PMC article.
References
- Aravin AA, Naumova NM, Tulin AV, Vagin VV, Rozovsky YM, Gvozdev VA. Double-stranded RNA-mediated silencing of genomic tandem repeats and transposable elements in the D. melanogaster germline. Curr Biol. 2001;11:1017–1027. - PubMed
- Aravin A, Gaidatzis D, Pfeffer S, Lagos-Quintana M, Landgraf P, Iovino N, Morris P, Brownstein MJ, Kuramochi-Miyagawa S, Nakano T, et al. A novel class of small RNAs bind to MILI protein in mouse testes. Nature. 2006;442:203–207. - PubMed
- Aravin AA, Hannon GJ, Brennecke J. The Piwi-piRNA pathway provides an adaptive defense in the transposon arms race. Science. 2007a;318:761–764. - PubMed
- Aravin AA, Sachidanandam R, Girard A, Fejes-Toth K, Hannon GJ. Developmentally regulated piRNA clusters implicate MILI in transposon control. Science. 2007b;316:744–747. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous