Control of autophagy initiation by phosphoinositide 3-phosphatase Jumpy - PubMed (original) (raw)
Control of autophagy initiation by phosphoinositide 3-phosphatase Jumpy
Isabelle Vergne et al. EMBO J. 2009.
Abstract
The majority of studies on autophagy, a cytoplasmic homeostasis pathway of broad biological and medical significance, have been hitherto focused on the phosphatidylinositol 3-kinases as the regulators of autophagy. Here, we addressed the reverse process driven by phosphoinositide phosphatases and uncovered a key negative regulatory role in autophagy of a phosphatidylinositol 3-phosphate (PI3P) phosphatase Jumpy (MTMR14). Jumpy associated with autophagic isolation membranes and early autophagosomes, defined by the key factor Atg16 necessary for proper localization and development of autophagic organelles. Jumpy orchestrated orderly succession of Atg factors by controlling recruitment to autophagic membranes of the sole mammalian Atg factor that interacts with PI3P, WIPI-1 (Atg18), and by affecting the distribution of Atg9 and LC3, the two Atg factors controlling organization and growth of autophagic membranes. A catalytically inactive Jumpy mutant, R336Q, found in congenital disease centronuclear myopathy, lost the ability to negatively regulate autophagy. This work reports for the first time that initiation of autophagy is controlled not only by the forward reaction of generating PI3P through a lipid kinase but that its levels are controlled by a specific PI3P phosphatase, which when defective can lead to human disease.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures
Figure 1
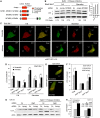
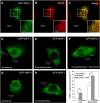
Screening of active members of the myotubularin family identifies Jumpy as a negative regulator of autophagy. (A) Domains and members of catalytically active myotubularins. Asterisks, Jumpy mutations found in patients with centronuclear myopathy. C330S, R336Q and Y462C, Jumpy mutants used in this study. (B) RAW 264.7 cells transfected for 48 h with control (sc), MTMR6 or Jumpy siRNA, were pretreated for 30 min with 100 nM Bafilomycin A1 (BafA1), 10 μg/ml E64d and 10 μg/ml pepstatin, then incubated for 30 min in full or starvation media in the presence of BafA1, E64d and pepstatin. Cells were lysed and analysed by immunoblotting with anti-LC3 or anti-actin. Densitometric LC3-II/actin ratios are shown underneath the blot. Graph: LC3-II/actin ratio for Jumpy siRNA in full medium is equal to or exceeds LC3-II/actin ratio for control siRNA (sc) in starvation medium. (C, D) RAW 264.7 cells were transfected for 36 h with control (scramble) or Jumpy siRNA, transfected once more with corresponding siRNA and mRFP-GFP-LC3 DNA construct overnight and incubated for 2 h in full or starvation media. Cells were fixed and LC3 puncta analysed by confocal fluorescence microscopy. (C) Representative confocal images of RAW 264.7 cells in full or starvation media after transfection with control (scramble) or Jumpy siRNA (siJumpy). Red and yellow arrows indicate GFP−RFP+ and GFP+RFP+ puncta, respectively. (D) Quantitation of number of LC3 puncta per cell, total puncta per cell (GFP+RFP+ and GFP−RFP+ puncta) GFP+RFP+ puncta per cell and GFP−RFP+ puncta per cell. Data, mean±s.e.m. _n_=5 independent experiments, 30 cells per experiments. *P<0.05, **P<0.01, ***P<0.001 (_t_-test). Scale bars, 5 μm. (E, F) C2C12 myoblasts were transfected for 48 h with control (scramble) or Jumpy siRNA, followed by a second transfection overnight with corresponding siRNA and mRFP-GFP-LC3 DNA construct. Cells were fixed and LC3 puncta were counted by confocal fluorescence microscopy. (E) Representative confocal images of C2C12 cells in full media after transfection with control (scramble) or Jumpy siRNA (siJumpy). (F) Quantification of the number of LC3 puncta per cell. Data, mean±SEM for _n_=3 (independent experiments), 30 cells per experiments. *P <0.05 (_t_-test). Scale bars, 10 μm. (G, H) C2C12 cells were transfected for 48 h with control (sc) (lanes 1, 2, 5, 6) or Atg5 siRNA (lanes 3, 4, 6, 7) followed by a second transfection for 48 h with same siRNA and control (lanes 1, 3, 5, 7) or Jumpy siRNA (lanes 2, 4, 6, 8). Cells were incubated for 1 h with or without 100 nM Baf A1, 10 μg/ml E64d and 10 μg/ml pepstatin in full media, lysed and analysed by immunoblotting with anti-LC3 or anti-actin (G). (H) Densitometric LC3-II/actin ratios for samples treated with BafA1 and protease inhibitors from G (lanes 5–8). Inset shows Atg5 knock-down by immunoblotting.
Figure 2
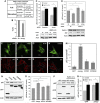
Jumpy blocks autophagic degradation. (A) Proteolysis of long-lived proteins in C2C12 myoblasts. C2C12 cells were transfected with control (scramble) or Jumpy siRNA (siJumpy), labelled overnight in media containing [3H] leucine, washed, incubated for 2 h in complete media (containing cold leucine) and incubated for 4 h in full or starvation media. Leucine release was calculated from radioactivity in the tricarboxylic acid-soluble form relative to total cell radioactivity. Results shown represent mean±s.e.m. for combined data from three independent experiments. (B) Quantitation of p62 protein levels. C2C12 cells were transfected for 48 h with control (sc) or Jumpy siRNA and p62 levels analysed by immunoblotting, quantitated by densitometry and represented as a percentage of control. Data are mean±s.e.m. (_n_=4 independent experiments). (C) C2C12 cells were transfected for 48 h with control (sc) or Jumpy siRNA, incubated for 4 h with or without 100 nM Bafilomycin A1 in full media, lysed, probed for endogenous p62 and actin by immunoblotting and percentage of p62 were quantitated (mean±s.e.m., _n_=3). (D) C2C12 cells were transfected for 48 h with control (sc) (lanes 1, 2) or Beclin siRNA (lanes 3, 4) followed by a second transfection for 48 h with same siRNA and control (lanes 1, 3) or Jumpy siRNA (lanes 2, 4). Cells were lysed, probed for p62, Beclin and actin by immunoblotting and percentage of p62 were quantitated (mean±s.e.m., _n_=5). (E–M) C2C12 cells were transfected for 48 h with GFP, GFP-Jumpy (Jumpy), GFP-MTM1 (MTM1) or GFP-MTMR2 (MTMR2), fixed and immunostained with anti-p62 antibody (red). p62 puncta were quantitated by confocal fluorescence microscopy. Representative confocal images of GFP (E), GFP-Jumpy (F), GFP-MTM1 (G), GFP-MTMR2 (H) transfected cells, immunostained for p62 (I), (J), (K) and (L), respectively. Scale bars, 5 μm. White lines represent outline of the cells. Quantitation of p62 puncta per cell (M). Bars, SEM (_n_=3 independent experiments, with an average of 30 cells per experiments). (N, O) C2C12 cells were transfected for 48 h with GFP, GFP-Jumpy (Jumpy), GFP-MTM1 (MTM1) or GFP-MTMR2 (MTMR2), lysed, analysed for p62, GFP and actin by immunoblotting (N) and percentage of p62 were quantitated (mean±s.e.m., _n_=3) (O). (P, Q) C2C12 cells were transfected for 48 h with GFP or GFP-Jumpy (Jumpy), incubated with or without 100 nM Bafilomycin A1 (BafA1) for 4 h, lysed, analysed for p62, GFP and actin by immunoblotting (P) and percentage of p62 were quantitated (mean±s.e.m., _n_=4) (Q). *P<0.05, ***P<0.001, †ns (_t_-test).
Figure 3
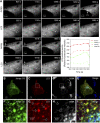
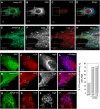
Jumpy localizes to autophagosomes. (A) Transient association of Jumpy WT with LC3+ organelles. Time lapse sequence of C2C12 myoblasts transfected for 24 h with GFP-Jumpy WT and Cherry-LC3 and analysed by live confocal microscopy in a 5LIVE Zeiss microscope. EBSS was added to the cells and _z_-stacks collected at 3-min intervals for a total of 45 min. The collected images were processed to generate a maximum projection (collapsing a 3D image into an x–y projection) for each time point. Arrows indicate colocalization of GFP-Jumpy WT with LC3. Graph: Plots of relative fluorescence intensity (GFP fluorescence pixel intensity) over time of GFP-Jumpy WT colocalizing with Cherry-LC3+-positive puncta indicated by arrows. (B–I) Association of Jumpy CS mutant with autophagic organelles. C2C12 cells were transfected for 24 h with GFP-Jumpy C330S (Jumpy CS) and tdTomato-LC3 (LC3), fixed and immunostained with anti-G58K (Alexa 648-labelled secondary antibody). Jumpy CS (green), LC3 (red) and G58K (white in D, H or blue in E, I). Boxed areas (B–E) are shown at higher magnification in the corresponding panel below (F–I). Scale bars, 2 μm. White arrows indicate colocalization between Jumpy CS, LC3 but not G58K, blue arrow indicates colocalization between Jumpy CS and G58K but not LC3.
Figure 4
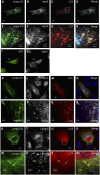
Jumpy localizes to autophagic isolation membranes. (A–H) C2C12 cells were transfected for 24 h with GFP-Jumpy C330S (Jumpy CS) and tdTomato-LC3 (LC3), fixed and immunostained with anti-Atg16 (Alexa 648-labelled secondary antibody). Jumpy CS (green), LC3 (red) and Atg16 (white in B and F or blue in D and H). Boxed areas (A–D) are shown at higher magnification in the corresponding panel below (E–H). White arrows indicate colocalization among Jumpy CS, LC3 and Atg16, yellow arrow indicates colocalization between Jumpy CS and LC3 but not Atg16, blue arrow indicates colocalization between Jumpy CS and Atg16 but not LC3. (I, J) C2C12 cells were transfected for 24 h with GFP (I), fixed and immunostained with anti-Atg16 (J). (K–R) C2C12 cells were transfected for 24 h with GFP-Jumpy C330S (Jumpy CS) and tdTomato-LC3 (LC3), fixed and immunostained with anti-Atg12(M) antibody. Jumpy CS (green), LC3 (red) and Atg12 (white in L, P or blue in N, R). Boxed areas (K–N) are shown at higher magnification in the corresponding panel below (O–R). White arrows indicate colocalization among Jumpy CS, LC3 and Atg12, yellow arrow indicates colocalization between Jumpy CS and LC3 but not Atg12, blue arrow indicates colocalization between Jumpy CS and Atg12 but not LC3. (S–ZA) C2C12 cells were transfected for 24 h with Jumpy CS and Cherry-LC3 (LC3), fixed and immunostained with anti-lamp1. Jumpy CS (green), LC3 (red) and Lamp1 (white in T, X or blue in V, ZA). Boxed areas (S–V) are shown at higher magnification in the corresponding panel below (W–ZA). Yellow arrows indicate colocalization between Jumpy CS and LC3 but not Lamp1. Scale bars, 2 μm.
Figure 5
Jumpy siRNA increases recruitment of PI3P-binding Atg factor WIPI-1 (Atg18) to autophagic membranes. (A–C) C2C12 were transfected for 48 h with Jumpy siRNA, followed by a second transfection overnight with Jumpy siRNA and GFP-WIPI-1, incubated for 4 h in starvation media, fixed and immunostained with anti-Atg16 antibody (red). GFP-WIPI1-transfected cell (A), endogenous Atg16 immunostaining (B), colocalization of GFP-WIPI-1 and Atg16 is indicated in yellow in merge image (C). Boxed areas are shown at higher magnification as inset. Scale bars, 10 and 5 μm in insets. (D–I) C2C12 cells were transfected for 48 h with control (scramble) (D, E) or Jumpy siRNA (F–H), followed by a second transfection overnight with corresponding siRNA and GFP-WIPI-1. Cells were incubated for 4 h in full (D, G) or starvation media (E, F, H) in presence (F) or absence (E–H) of 100 nM wortmannin, fixed and analysed by confocal fluorescence microscopy. Quantitation of WIPI-1 puncta per cell (I). Bars, s.e.m. (_n_=3 independent experiments, with an average of 30 cells per experiment). *P<0.05, **P<0.01 (_t_-test).
Figure 6
Jumpy colocalizes with and regulates Atg9 distribution to autophagic organelles. (A–T) C2C12 cells were transfected for 24 h with GFP, GFP-Jumpy WT or GFP-Jumpy C330S (Jumpy CS) and Tomato-LC3 (LC3) and HA-Atg9, fixed and immunostained with anti-HA. GFP (I), Jumpy WT (M), Jumpy CS (green) (A, E, Q), LC3 (red) (C, G, J, N, R) and Atg9 (white in B, F, K, O, S or blue in D, H, I, P, T). Boxed areas (A–D) are shown at higher magnification in the corresponding panel below (E–H). White arrows indicate colocalization between Jumpy CS, LC3 and Atg9, blue arrow indicates colocalization between Jumpy CS and Atg9 but not LC3 and yellow arrow indicates colocalization between Jumpy CS and LC3 but not Atg9. White boxes (I–T) show LC3 puncta location. Scale bars, 5 μm. (U) Percentage of LC3 puncta colocalizing with Atg9. n, number of puncta counted.
Figure 7
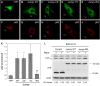
Jumpy R336Q mutant associated with centronuclear myopathy is defective in inhibiting autophagy. (A–K) C2C12 cells were transfected for 48 h with YFP, YFP-Jumpy WT (Jumpy WT), YFP-Jumpy C330S (Jumpy CS), YFP-Jumpy Y462C (Jumpy YC) or YFP-Jumpy R336Q (Jumpy RQ), fixed and immunostained with anti-p62 antibody (red). p62 puncta were quantitated by confocal fluorescence microscopy. Representative confocal images of YFP (A), YFP-Jumpy WT (B), YFP-Jumpy C330S (C), YFP-Jumpy Y462C (D) and YFP-Jumpy R336Q (E) transfected cells, immunostained for p62 (F), (G), (H), (I) and (J), respectively. Scale bars, 5 μm. Quantitation of p62 puncta per cell (K). Bars, s.e.m. (_n_=3 independent experiments, 30 cells per experiments). *P<0.05 (_t_-test), ns: nonsignificant. (L) HeLa cells were transfected for 24 h with YFP, YFP-Jumpy WT (Jumpy WT) or YFP-Jumpy R336Q (Jumpy RQ), incubated with or without 50 ng/μl rapamycin (BafA1 (100 nM) was present in both control and rapamycin treated cells) for 2 h, lysed and analysed for LC3, YFP and actin by immunoblotting. Densitometric LC3-II/actin ratios are shown underneath the blot.
Similar articles
- A novel PtdIns3P and PtdIns(3,5)P2 phosphatase with an inactivating variant in centronuclear myopathy.
Tosch V, Rohde HM, Tronchère H, Zanoteli E, Monroy N, Kretz C, Dondaine N, Payrastre B, Mandel JL, Laporte J. Tosch V, et al. Hum Mol Genet. 2006 Nov 1;15(21):3098-106. doi: 10.1093/hmg/ddl250. Epub 2006 Sep 28. Hum Mol Genet. 2006. PMID: 17008356 - Primary T-tubule and autophagy defects in the phosphoinositide phosphatase Jumpy/MTMR14 knockout mice muscle.
Hnia K, Kretz C, Amoasii L, Böhm J, Liu X, Messaddeq N, Qu CK, Laporte J. Hnia K, et al. Adv Biol Regul. 2012 Jan;52(1):98-107. doi: 10.1016/j.advenzreg.2011.09.007. Adv Biol Regul. 2012. PMID: 21930146 No abstract available. - WIPI-Mediated Autophagy and Longevity.
Grimmel M, Backhaus C, Proikas-Cezanne T. Grimmel M, et al. Cells. 2015 May 22;4(2):202-17. doi: 10.3390/cells4020202. Cells. 2015. PMID: 26010754 Free PMC article. Review. - The role of PI3P phosphatases in the regulation of autophagy.
Vergne I, Deretic V. Vergne I, et al. FEBS Lett. 2010 Apr 2;584(7):1313-8. doi: 10.1016/j.febslet.2010.02.054. Epub 2010 Feb 24. FEBS Lett. 2010. PMID: 20188094 Free PMC article. Review. - Regulation of membrane biogenesis in autophagy via PI3P dynamics.
Noda T, Matsunaga K, Taguchi-Atarashi N, Yoshimori T. Noda T, et al. Semin Cell Dev Biol. 2010 Sep;21(7):671-6. doi: 10.1016/j.semcdb.2010.04.002. Epub 2010 Apr 18. Semin Cell Dev Biol. 2010. PMID: 20403452 Review.
Cited by
- The beneficial role of proteolysis in skeletal muscle growth and stress adaptation.
Bell RA, Al-Khalaf M, Megeney LA. Bell RA, et al. Skelet Muscle. 2016 Apr 6;6:16. doi: 10.1186/s13395-016-0086-6. eCollection 2016. Skelet Muscle. 2016. PMID: 27054028 Free PMC article. Review. - PI(5)P regulates autophagosome biogenesis.
Vicinanza M, Korolchuk VI, Ashkenazi A, Puri C, Menzies FM, Clarke JH, Rubinsztein DC. Vicinanza M, et al. Mol Cell. 2015 Jan 22;57(2):219-34. doi: 10.1016/j.molcel.2014.12.007. Epub 2015 Jan 8. Mol Cell. 2015. PMID: 25578879 Free PMC article. - PtdIns(4,5)P₂ and PtdIns3P coordinate to regulate phagosomal sealing for apoptotic cell clearance.
Cheng S, Wang K, Zou W, Miao R, Huang Y, Wang H, Wang X. Cheng S, et al. J Cell Biol. 2015 Aug 3;210(3):485-502. doi: 10.1083/jcb.201501038. J Cell Biol. 2015. PMID: 26240185 Free PMC article. - The role of lipids in autophagy and its implication in neurodegeneration.
Hernandez-Diaz S, Soukup SF. Hernandez-Diaz S, et al. Cell Stress. 2020 May 19;4(7):167-186. doi: 10.15698/cst2020.07.225. Cell Stress. 2020. PMID: 32656499 Free PMC article. Review. - Cellular and molecular mechanisms of muscle atrophy.
Bonaldo P, Sandri M. Bonaldo P, et al. Dis Model Mech. 2013 Jan;6(1):25-39. doi: 10.1242/dmm.010389. Dis Model Mech. 2013. PMID: 23268536 Free PMC article. Review.
References
- Alonso A, Sasin J, Bottini N, Friedberg I, Friedberg I, Osterman A, Godzik A, Hunter T, Dixon J, Mustelin T (2004) Protein tyrosine phosphatases in the human genome. Cell 117: 699–711 - PubMed
- Bitoun M, Maugenre S, Jeannet PY, Lacene E, Ferrer X, Laforet P, Martin JJ, Laporte J, Lochmuller H, Beggs AH, Fardeau M, Eymard B, Romero NB, Guicheney P (2005) Mutations in dynamin 2 cause dominant centronuclear myopathy. Nat Genet 37: 1207–1209 - PubMed
- Blondeau F, Laporte J, Bodin S, Superti-Furga G, Payrastre B, Mandel JL (2000) Myotubularin, a phosphatase deficient in myotubular myopathy, acts on phosphatidylinositol 3-kinase and phosphatidylinositol 3-phosphate pathway. Hum Mol Genet 9: 2223–2229 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases