Docking of a specialized PIP Box onto chromatin-bound PCNA creates a degron for the ubiquitin ligase CRL4Cdt2 - PubMed (original) (raw)
Docking of a specialized PIP Box onto chromatin-bound PCNA creates a degron for the ubiquitin ligase CRL4Cdt2
Courtney G Havens et al. Mol Cell. 2009.
Abstract
Substrates of the E3 ubiquitin ligase CRL4(Cdt2), including Cdt1 and p21, contain a PCNA-binding motif called a PIP box. Upon binding of the PIP box to PCNA on chromatin, CRL4(Cdt2) is recruited and the substrate is ubiquitylated. Importantly, a PIP box cannot be sufficient for destruction, as most PIP box proteins are stable. Using Xenopus egg extracts, we identify two sequence elements in CRL4(Cdt2) substrates that promote their proteolysis: a specialized PIP box that confers exceptionally efficient PCNA binding and a basic amino acid 4 residues downstream of the PIP box, which recruits CRL4(Cdt2) to the substrate-PCNA complex. We also identify two mechanisms that couple CRL4(Cdt2)-dependent proteolysis to the chromatin-bound form of PCNA, ensuring that this proteolysis pathway is active only in S phase or after DNA damage. Thus, CRL4(Cdt2) recognizes an unusual degron, which is assembled specifically on chromatin via the binding of a specialized PIP box to PCNA.
Figures
Figure 1. Cdt11-243 is destroyed in a CRL4Cdt2- and PCNA-dependent manner and interacts with chromatin-bound PCNA
(A) Model of PCNA-Cdt1-CRL4Cdt2 complex on DNA (B) Schematic of PCNA binding and licensing regions of Xenopus Cdt1. (C) Cdt2-dependent destruction of endogenous Cdt1 and recombinant Cdt11-243 in HSS. Mock-depleted or Cdt2-depleted HSS was supplemented with 50 nM recombinant Cdt11-243-3xNLS-GST-Flag (Cdt11-243) and 5 ng/μl MMS plasmid. At different times, samples were blotted for the indicated proteins. Asterisks indicate background bands. (D) Cdt11-243 destruction is PIP box-dependent. HSS was supplemented with 1 kb MMS DNA and Cdt11-243 or Cdt11-243/ΔPIP. At different times, samples were blotted for the indicated proteins. The sequence of the ΔPIP mutant is shown. (E) Cdt1-depleted HSS was supplemented with immobilized, 1 kb MMS DNA and 2 mg/ml methyl ubiquitin. 50 nM Cdt11-243 or Cdt11-243/ΔPIP was added, as indicated, and after 10 minutes, chromatin was recovered from the extract, washed, and blotted for the indicated proteins.
Figure 2. Specific assembly of the PCNA-Cdt1-CRL4Cdt2 complex on chromatin
(A) 10 μl aliquots of HSS supplemented with methyl ubiquitin were mixed with 350 ng of 1 kb MMS DNA (lanes 1-3), no DNA (lanes 4-6), or 350 ng immobilized 1 kb MMS DNA template (lanes 7-9), as well as 250 nM Cdt11-243-Flag or Cdt11-243/ΔPIP-Flag for 10 minutes. For lanes 1-6, Cdt11-243 was precipitated with flag antibody and the IPs were analyzed. For lanes 7-10, the beads were recovered and associated proteins analyzed. In lanes 11-13, input extract was analyzed. The material recovered from the equivalent of 2 μl of HSS was loaded in lanes 1-10, whereas 1 μl of HSS was loaded in lanes 11-13. HC, heavy chain; LC, light chain. (B) For reactions described in lanes 1-3 of Panel (A), DNA was extracted from the Flag IP, 50% was analyzed on an agarose gel and then stained with SYBER gold alongside the total input DNA, which was also extracted, but not exposed to extract. (C) 10 μl aliquots of HSS supplemented with methyl ubiquitin were mixed with 350 ng of 1 kb MMS DNA (lanes 1-3) or 350 ng immobilized 1 kb MMS DNA template (lanes 7-9), as well as 250 nM Flag-Fen1 or Flag-Fen1ΔPIP for 10 minutes. For lanes 1-6, Flag-Fen1 was precipitated with flag antibody and the IPs were analyzed. For lanes 7-10, the DNA was recovered and associated proteins analyzed. In lanes 11-13, input extract was analyzed. The material recovered from the equivalent of 2 μl of HSS was loaded in lanes 1-10, whereas 0.5 μl of HSS was loaded in lanes 11-13.
Figure 3. The p21 PIP box peptide is sufficient to recruit CRL4Cdt2 to chromatin
(A) Sequence of human p21 PIP box peptides used in panels (B) and (C) aligned with Xenopus Cdt1. (B) HSS was mixed with methyl ubiquitin and immobilized 1 kb MMS DNA. Three minutes after the addition of DNA, 10 μM p21ΔPIP peptide (MUT), p21 peptide (WT), or p21R+4A peptide (R+4A) were added to the reaction. Samples were stopped with sample buffer after 10 minutes and blotted for the indicated proteins. (C) LSS was supplemented with methyl ubiquitin, sperm chromatin and buffer, p21ΔPIP peptide (Mut) or p21 peptide (WT). After 45 minutes, chromatin was isolated, washed, and blotted for the indicated proteins. All samples were run on the same gel, but some irrelevant lanes were removed between lanes 2 and 3.
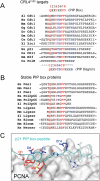
Figure 4. Sequence alignment of PIP box proteins
(A) Alignment of PIP boxes from proteins that are known to be targeted for destruction by CRL4Cdt2. Canonical PIP box residues are shown in red and putative “degron-specific” residues are shown in blue. The PIP box consensus, and the putative PIP degron consensus are shown (ψ = I/L/M/V, J = Y/F, B=K/R). The absence of a dash indicates the N or C terminus. (B) Alignment of PIP boxes from proteins that are not likely targets of CRL4Cdt2. (C) An image of the PCNA-p21 peptide co-crystal structure (Gulbis et al., 1996) was generated using PDB accession number 1AXC and PyMOL (
).
Figure 5. Mutational analysis of the Cdt1 PIP box
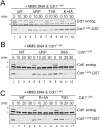
(A) HSS was mixed with MMS plasmid and 50 nM recombinant Cdt11-243, Cdt11-243/ΔPIP, Cdt11-243/T5A, or Cdt11-243/K+4A. Reactions were stopped at different times and blotted for Cdt1 or Cdt11-243-GST (anti-GST antibody). (B) HSS was mixed with MMS plasmid and 50 nM recombinant Cdt11-243, Cdt11-243/ΔPIP, or Cdt11-243/D6A. Reactions were stopped at different times and blotted as in (A). (C) HSS was mixed with MMS plasmid and 50 nM recombinant Cdt11-243, Cdt11-243/ΔPIP, Cdt11-243/R+5A or Cdt11-243/T5D. Reactions were stopped at the indicated times and blotted as in (A).
Figure 6. PCNAChromatin binding and Cdt2 recruitment of Cdt11-243 mutants
(A) Cdt1-depleted HSS was supplemented with immobilized 1 kb MMS DNA, methyl ubiquitin, and 50 nM Cdt11-243, Cdt11-243/ΔPIP, Cdt11-243/K+4A or Cdt11-243/T5A. After 10 minutes, the beads were recovered, washed with buffer containing different concentrations of salt, and blotted for the indicated proteins. (B) Cdt1-depleted HSS was mixed with immobilized 1 kb MMS DNA, methyl ubiquitin and 50 nM (“1x”) to 500 nM (“10x”) Cdt11-243 or Cdt11-243/K+4A. After 10 minutes, the beads were recovered and blotted for the indicated proteins. (C) Cdt1-depleted HSS was mixed with immobilized 1 kb MMS DNA, methyl ubiquitin, and 250 (5x) or 500 nM (10x) Cdt11-243, Cdt11-243/T5A, or Cdt11-243/T5D. Chromatin-bound proteins were analyzed as in (B). (D) Cdt1-depleted HSS was mixed with immobilized 1 kb MMS DNA, methyl ubiquitin, and 50 nM Cdt11-243 or Cdt11-243/D6A. Chromatin-bound proteins were analyzed as in (B). All samples were run on the same gel, but some irrelevant lanes were removed between lanes 2 and 3.
Figure 7. The PIP degron is portable
(A) Sequence comparison of Xenopus Cdt1's PIP box with that of Xenopus Fen1 and various mutants of Fen1. (B) HSS was mixed with MMS plasmid and 50 nM recombinant Fen1, Fen1T, or Fen1TK. Reactions were stopped at the indicated times and blotted for Cdt1 and the Flag peptide, to visualize Flag-Fen1. (C) Mock-depleted or Cdt2-depleted HSS was supplemented with 5 ng/μl MMS plasmid and 50 nM Fen1 or Fen1TK. Samples were blotted for the indicated proteins. (D) HSS was incubated with 50 μM mutant or WT p21 PIP peptide and then supplemented with 50 nM recombinant Fen1TK or Fen1 and 5 ng/μl MMS plasmid. At different times, samples were blotted for the indicated proteins. (E) HSS was mixed with immobilized 1 kb MMS DNA, methyl ubiquitin, and 200 nM Fen1, Fen1ΔPIP, Fen1T or Fen1TK. After 10 minutes, the beads were recovered and blotted for the indicated proteins. (F) 10 μl aliquots of HSS supplemented with methyl ubiquitin were mixed with 350 ng of 1 kb MMS DNA (lanes 1-3) or 350 ng immobilized 1 kb MMS DNA (lanes 7-9), as well as 250 nM Cdt11-243-Flag, Flag-Fen1T or Flag-Fen1TK for 10 minutes. Lane 10 contained all three proteins, HSS and magnetic beads, but no DNA. For lanes 1-6, Flag-tagged proteins were precipitated with flag antibody and the IPs were analyzed. For lanes 7-10, the beads were recovered and associated proteins analyzed. In lanes 11-13, input extract was analyzed. The material recovered from the equivalent of 2 μl of HSS was loaded in lanes 1-10, whereas 0.5 μl of HSS was loaded in lanes 11-13. All samples were from the same experiment, but for the anti-Flag panel, a darker exposure of lanes 7-13 is shown. Lower panel, coomassie gel showing that equal amounts of recombinant proteins were used. Note that Cdt11-243-Flag contains a single C-terminal Flag tag, whereas Flag-Fen1 contains 2 N-terminal Flag tags.
Similar articles
- Direct role for proliferating cell nuclear antigen in substrate recognition by the E3 ubiquitin ligase CRL4Cdt2.
Havens CG, Shobnam N, Guarino E, Centore RC, Zou L, Kearsey SE, Walter JC. Havens CG, et al. J Biol Chem. 2012 Mar 30;287(14):11410-21. doi: 10.1074/jbc.M111.337683. Epub 2012 Feb 2. J Biol Chem. 2012. PMID: 22303007 Free PMC article. - The CRL4Cdt2 ubiquitin ligase mediates the proteolysis of cyclin-dependent kinase inhibitor Xic1 through a direct association with PCNA.
Kim DH, Budhavarapu VN, Herrera CR, Nam HW, Kim YS, Yew PR. Kim DH, et al. Mol Cell Biol. 2010 Sep;30(17):4120-33. doi: 10.1128/MCB.01135-09. Epub 2010 Jul 6. Mol Cell Biol. 2010. PMID: 20606006 Free PMC article. - Thymine DNA glycosylase is a CRL4Cdt2 substrate.
Slenn TJ, Morris B, Havens CG, Freeman RM Jr, Takahashi TS, Walter JC. Slenn TJ, et al. J Biol Chem. 2014 Aug 15;289(33):23043-23055. doi: 10.1074/jbc.M114.574194. Epub 2014 Jun 19. J Biol Chem. 2014. PMID: 24947512 Free PMC article. - CRL4Cdt2 Ubiquitin Ligase, A Genome Caretaker Controlled by Cdt2 Binding to PCNA and DNA.
Mazian MA, Yamanishi K, Rahman MZA, Ganasen M, Nishitani H. Mazian MA, et al. Genes (Basel). 2022 Jan 29;13(2):266. doi: 10.3390/genes13020266. Genes (Basel). 2022. PMID: 35205311 Free PMC article. Review. - Regulation of DNA Replication Licensing and Re-Replication by Cdt1.
Zhang H. Zhang H. Int J Mol Sci. 2021 May 14;22(10):5195. doi: 10.3390/ijms22105195. Int J Mol Sci. 2021. PMID: 34068957 Free PMC article. Review.
Cited by
- The Many Roles of PCNA in Eukaryotic DNA Replication.
Boehm EM, Gildenberg MS, Washington MT. Boehm EM, et al. Enzymes. 2016;39:231-54. doi: 10.1016/bs.enz.2016.03.003. Epub 2016 Apr 19. Enzymes. 2016. PMID: 27241932 Free PMC article. Review. - CRL4(CDT2) targets CHK1 for PCNA-independent destruction.
Huh J, Piwnica-Worms H. Huh J, et al. Mol Cell Biol. 2013 Jan;33(2):213-26. doi: 10.1128/MCB.00847-12. Epub 2012 Oct 29. Mol Cell Biol. 2013. PMID: 23109433 Free PMC article. - Two different replication factor C proteins, Ctf18 and RFC1, separately control PCNA-CRL4Cdt2-mediated Cdt1 proteolysis during S phase and following UV irradiation.
Shiomi Y, Hayashi A, Ishii T, Shinmyozu K, Nakayama J, Sugasawa K, Nishitani H. Shiomi Y, et al. Mol Cell Biol. 2012 Jun;32(12):2279-88. doi: 10.1128/MCB.06506-11. Epub 2012 Apr 9. Mol Cell Biol. 2012. PMID: 22493068 Free PMC article. - PCNA-coupled p21 degradation after DNA damage: The exception that confirms the rule?
Soria G, Gottifredi V. Soria G, et al. DNA Repair (Amst). 2010 Apr 4;9(4):358-64. doi: 10.1016/j.dnarep.2009.12.003. Epub 2010 Jan 8. DNA Repair (Amst). 2010. PMID: 20060369 Free PMC article. Review. - DDB2 (damaged-DNA binding protein 2) in nucleotide excision repair and DNA damage response.
Stoyanova T, Roy N, Kopanja D, Raychaudhuri P, Bagchi S. Stoyanova T, et al. Cell Cycle. 2009 Dec 15;8(24):4067-71. doi: 10.4161/cc.8.24.10109. Epub 2009 Dec 17. Cell Cycle. 2009. PMID: 19923893 Free PMC article. Review.
References
- Ang XL, Wade Harper J. SCF-mediated protein degradation and cell cycle control. Oncogene. 2005;24:2860–2870. - PubMed
- Arias EE, Walter JC. PCNA functions as a molecular platform to trigger Cdt1 destruction and prevent re-replication. Nat Cell Biol. 2006;8:84–90. - PubMed
- Chuang LC, Yew PR. Regulation of nuclear transport and degradation of the Xenopus cyclin-dependent kinase inhibitor, p27Xic1. J Biol Chem. 2001;276:1610–1617. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- F32 GM082014/GM/NIGMS NIH HHS/United States
- R01 GM062267-08/GM/NIGMS NIH HHS/United States
- R01 GM062267/GM/NIGMS NIH HHS/United States
- R01 GM080676/GM/NIGMS NIH HHS/United States
- R01 GM080676-02/GM/NIGMS NIH HHS/United States
- F32-GM082014/GM/NIGMS NIH HHS/United States
- R01-GM080676/GM/NIGMS NIH HHS/United States
- R01 GM062267-09/GM/NIGMS NIH HHS/United States
- R01 GM080676-01A1/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous