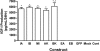The insulin-like growth factor (IGF)-I E-peptides modulate cell entry of the mature IGF-I protein - PubMed (original) (raw)
The insulin-like growth factor (IGF)-I E-peptides modulate cell entry of the mature IGF-I protein
Lindsay A Pfeffer et al. Mol Biol Cell. 2009 Sep.
Abstract
Insulin-like growth factor (IGF)-I is a critical protein for cell development and growth. Alternative splicing of the igf1 gene gives rise to multiple isoforms. In rodents, proIGF-IA and proIGF-IB have different carboxy-terminal extensions called the E-peptides (EA and EB) and upon further posttranslational processing, produce the identical mature IGF-I protein. Rodent EB has been reported to have mitogenic and motogenic effects independent of IGF-I. However, effects of EA or EB on mature IGF-I, or whether proIGF-IA and proIGF-IB have different properties, have not been addressed. To determine whether the presence of EA or EB affected the distribution and stability of mature IGF-I protein, transient transfections of cDNAs encoding murine IGF-IA, IGF-IB, and mature IGF-I were performed in C2C12 cells, a skeletal muscle cell line. IGF-I secretion was measured by enzyme-linked immunosorbent assay of the media, and did not differ between expression of proIGF-IA, proIGF-IB, or mature IGF-I expression. Next, epitope-tagged constructs were transfected to determine cellular distribution of IGF-I, EA, and EB in the cells throughout the culture. IGF-I was detected in significantly fewer nontransfected cells in cultures transfected with mature IGF-I compared with transfection of proIGF-IA or proIGF-IB. These results demonstrate that EA and EB are not required for IGF-I secretion but that they increase cell entry of IGF-I from the media. This study provides evidence that the EA and EB may modulate IGF-I in addition to having independent activity.
Figures
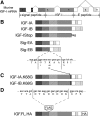
Figure 1.
Schematic of cDNA constructs generated for this study. (A) Exons and alternative splicing in murine Igf1. Alternative splicing of exons 1 or 2 to exon 3 produces the signal peptide. The mature IGF-I protein is invariant and encoded by exons 3 and 4. The C-terminal E-peptides are encoded by exons 4, 5, and 6, and alternative splicing of exon 5 inclusion gives rise to divergent E-peptides. (B) Constructs that retain or exclude mature IGF-I or the E-peptides. IGF-IA retains mature IGF-I and the EA-Peptide. IGF-IB retains mature IGF-I and the EB-peptide. IGF-IStop retains mature IGF-I but excludes both E-peptides. SigEA and SigEB exclude mature IGF-I but retain the EA- and EB-peptides, respectively. (C) Mutagenesis of mature IGF-I/E-peptide processing site. Lysine 68 was changed to glycine to block the primary cleavage site between the mature IGF-I protein and the E-peptides. (D) Addition of epitope tags to IGF-I constructs. A FLAG tag was inserted between the signal peptide and mature IGF-I immediately after the processing site. In separate constructs, an HA tag was added to the C terminus of the EA- or EB-peptide.
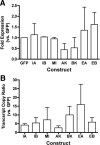
Figure 2.
Transfection and expression efficiency for IGF-I constructs. Data are presented as mean and SEs from three independent experiments. (A) Efficiency of transfection was determined in using the relative GFP expression compared with 18s as a housekeeping gene. Data are normalized to empty vector control that expresses only GFP. No statistical difference in transfection efficiency was found among any construct used in the study. (B) Efficiency of expression of the IGF-I cDNA insert for each plasmid was determined by the transcript copy ratio of the upstream IGF-I insert to the downstream GFP insert. Transcript copies were calculated from standard curves generated for each plasmid using the primer pairs listed in Table 1. Constructs GFP (empty vector control), IA (IGF-IA), IB, (IGF-IB), MI (Mature IGF-I/IGF-Istop), AK (IGF-IAK68G), BK (IGF-IBK68G), EA (SigEA), and EB (SigEB).
Figure 3.
IGF-I production is not affected by the presence of the E-peptides. Media content of IGF-I served as an index of IGF-I production after transfection. Production of IGF-I was significantly higher than control cells when the cDNA construct contained the sequence encoding mature IGF-I (IGF-IA, IGF-IB, IGF-IStop, IGF-IA.K68G [AK68G], and IGF-IB.K68G [BK68G]). No change in IGF-I production was afforded by the presence of the E-peptides (IGF-IA or IGF-IB compared with IGF-IStop; SigEA or SigEB compared with control). IGF-IB.K68G had higher IGF-I production than IGF-IA.K68G. The transfection conditions (mock) or the vector alone (GFP) did not affect IGF-I production. Levels of IGF-I production from Control, Mock, GFP, EA, and EB ranged from 4 to 10 pg/ml and are not apparent on the graph. Statistical comparisons from Tukey's multiple comparisons test; *p < 0.05 for comparison to control cultures; †p < 0.05 for comparisons between isoforms. Constructs: IA (IGF-IA), IB, (IGF-IB), MI (Mature IGF-I/IGF-Istop), AK (IGF-IAK68G), BK (IGF-IBK68G), EA (SigEA), EB (SigEB), GFP (empty vector control), Mock (Lipofectamine only), and Cont (no transfection).
Figure 4.
Form of secreted IGF-I from C2C12 cells after transfection. Immunoblotting of concentrated media with anti-FLAG was used to distinguish between proIGF-I and mature IGF-I in the culture media (left). Both pro- (bands a and b) and fully processed (mature, band e) IGF-I were detected when IGF-IA (IA) or IGF-IB (IB) constructs were transfected. IGF-IA lanes had a higher molecular weight band (a) consistent with glycosylated proIGF-IA, shown in the right panel. IGF-IB lanes had an additional lower molecular weight band (c) that could result from protease cleavage within the EB-peptide (right). Mature IGF-I (e) could be produced by IGF-IStop (ISt), and this lane serves as a control for the size of secreted IGF-I. However, a higher molecular weight band occurred (band d), which was not evident in the IGF-IA and IGF-IB lanes. NT, media from nontransfected cultures.
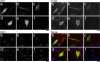
Figure 5.
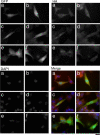
Cellular distribution of FLAG epitope-tagged IGF-I constructs FLAG-IGF-IA (a), FLAG-IGF-IB (b), FLAG-IGF-Istop (c), FLAG-IGF-IAK68G (d), FLAG-IGF-IBK68G (e), and no transfection (f). GFP serves as an indicator of positive transfection and is found in the cytoplasm and nucleus of all transfected cells. FLAG is detected in both transfected and nontransfected cells. It is found throughout the cytoplasm and concentrated in the perinuclear regions in transfected and nontransfected cells. Cleavage mutant constructs (d and e) have altered cell shape and lower intensity FLAG staining in the multiple cytoplasmic extensions. DAPI staining identifies the number of nuclei within each field. The merged images are pseudocolored, with GFP as green, FLAG as red, and DAPI as blue. Bar, 10 μm.
Figure 6.
Cellular distribution of HA epitope-tagged IGF-I constructs HA-IGF-IA (a), HA-IGF-IB (b), HA-SigEA (c), HA-SigEB (d), HA-IGF-IAK68G (e), and HA-IGF-IBK68G (f). GFP serves as an indicator of positive transfection and is found in the cytoplasm and nucleus of all transfected cells, as in Figure 5. HA is detected in both transfected and nontransfected cells. It is found throughout the cytoplasm and concentrated in the perinuclear regions in transfected and nontransfected cells. Cleavage mutant constructs (e and f) have altered cell shape and little detectable HA staining in the multiple cytoplasmic extensions. DAPI staining identifies the number of nuclei within each field. The merged images are pseudocolored, with GFP as green, HA as red, and DAPI as blue. Bar, 10 μm.
Figure 7.
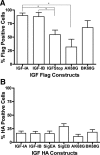
Proportion of C2C12 cells that internalize epitope tags after transfection. (A) FLAG uptake is dependent upon the transfected IGF-I construct. There was no difference in the proportion of FLAG-positive cells after transfection of FLAG-IGF-IA or FLAG-IGF-IB. Expression of mature IGF-I (FLAG-IGF-IStop) significantly reduced the proportion of FLAG-positive cells. Mutation of the primary cleavage site between mature IGF and the EA-peptide in IGF-IA (AK68G) also resulted in a significant decrease in FLAG-positive cells. (B) HA uptake is independent of the transfected IGF construct. There was no significant difference in the proportion of HA-positive cells in the presence or absence of the sequence encoding mature IGF. Mutation of the primary cleavage sites between IGF and the E-peptide (AK68G, BK68G) did not affect the proportion of HA-positive cells. Comparisons by one-way ANOVA followed by Tukey's multiple comparison test.
Similar articles
- Monoclonal antibodies to the carboxy-terminal Ea sequence of pro-insulin-like growth factor-IA (proIGF-IA) recognize proIGF-IA secreted by IM9 B-lymphocytes.
Wilson HE, Westwood M, White A, Clayton PE. Wilson HE, et al. Growth Horm IGF Res. 2001 Feb;11(1):10-7. doi: 10.1054/ghir.2000.0182. Growth Horm IGF Res. 2001. PMID: 11437469 - Human IGF-I propeptide A promotes articular chondrocyte biosynthesis and employs glycosylation-dependent heparin binding.
Shi S, Kelly BJ, Wang C, Klingler K, Chan A, Eckert GJ, Trippel SB. Shi S, et al. Biochim Biophys Acta Gen Subj. 2018 Mar;1862(3):567-575. doi: 10.1016/j.bbagen.2017.11.017. Epub 2017 Nov 21. Biochim Biophys Acta Gen Subj. 2018. PMID: 29174671 Free PMC article. - Insulin-like growth factor-I E-peptide activity is dependent on the IGF-I receptor.
Brisson BK, Barton ER. Brisson BK, et al. PLoS One. 2012;7(9):e45588. doi: 10.1371/journal.pone.0045588. Epub 2012 Sep 21. PLoS One. 2012. PMID: 23029120 Free PMC article. - The complexity of the IGF1 gene splicing, posttranslational modification and bioactivity.
Philippou A, Maridaki M, Pneumaticos S, Koutsilieris M. Philippou A, et al. Mol Med. 2014 May 7;20(1):202-14. doi: 10.2119/molmed.2014.00011. Mol Med. 2014. PMID: 24637928 Free PMC article. Review. - Role of Alternatively Spliced Messenger RNA (mRNA) Isoforms of the Insulin-Like Growth Factor 1 (IGF1) in Selected Human Tumors.
Kasprzak A, Szaflarski W. Kasprzak A, et al. Int J Mol Sci. 2020 Sep 23;21(19):6995. doi: 10.3390/ijms21196995. Int J Mol Sci. 2020. PMID: 32977489 Free PMC article. Review.
Cited by
- IGF expression in HPV-related and HPV-unrelated human cancer cells.
Durzyńska J, Barton E. Durzyńska J, et al. Oncol Rep. 2014 Sep;32(3):893-900. doi: 10.3892/or.2014.3329. Epub 2014 Jul 11. Oncol Rep. 2014. PMID: 25018100 Free PMC article. - New Modulators for IGF-I Activity within IGF-I Processing Products.
Brisson BK, Barton ER. Brisson BK, et al. Front Endocrinol (Lausanne). 2013 Mar 27;4:42. doi: 10.3389/fendo.2013.00042. eCollection 2013. Front Endocrinol (Lausanne). 2013. PMID: 23543904 Free PMC article. - The pro-forms of insulin-like growth factor I (IGF-I) are predominant in skeletal muscle and alter IGF-I receptor activation.
Durzyńska J, Philippou A, Brisson BK, Nguyen-McCarty M, Barton ER. Durzyńska J, et al. Endocrinology. 2013 Mar;154(3):1215-24. doi: 10.1210/en.2012-1992. Epub 2013 Feb 13. Endocrinology. 2013. PMID: 23407451 Free PMC article. - MUC4 is negatively regulated through the Wnt/β-catenin pathway via the Notch effector Hath1 in colorectal cancer.
Pai P, Rachagani S, Dhawan P, Sheinin YM, Macha MA, Qazi AK, Chugh S, Ponnusamy MP, Mallya K, Pothuraju R, Batra SK. Pai P, et al. Genes Cancer. 2016 May;7(5-6):154-168. doi: 10.18632/genesandcancer.108. Genes Cancer. 2016. PMID: 27551331 Free PMC article. - Deletion of muscle GRP94 impairs both muscle and body growth by inhibiting local IGF production.
Barton ER, Park S, James JK, Makarewich CA, Philippou A, Eletto D, Lei H, Brisson B, Ostrovsky O, Li Z, Argon Y. Barton ER, et al. FASEB J. 2012 Sep;26(9):3691-702. doi: 10.1096/fj.11-203026. Epub 2012 May 30. FASEB J. 2012. PMID: 22649033 Free PMC article.
References
- Adamo M. L., Neuenschwander S., LeRoith D., Roberts C. T. Structure, expression, and regulation of the IGF-1 gene. Adv. Exp. Med. Biol. 1993;343:1–11. - PubMed
- Adams G. R., McCue S. A. Localized infusion of IGF-I results in skeletal muscle hypertrophy in rats. J. Appl. Physiol. 1998;84:1716–1722. - PubMed
- Bach M. A., Roberts C. T., Jr., Smith E. P., LeRoith D. Alternative splicing produces messenger RNAs encoding insulin-like growth factor-I prohormones that are differentially glycosylated in vitro. Mol. Endocrinol. 1990;4:899–904. - PubMed
- Barr P. J. Mammalian subtilisins: the long-sought dibasic processing endoproteases. Cell. 1991;66:1–3. - PubMed
- Barton E. R. The ABCs of IGF-I isoforms: impact on muscle hypertrophy and implications for repair. Appl. Physiol. Nutr. Metab. 2006a;31:791–797. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous