Bcl6 and Blimp-1 are reciprocal and antagonistic regulators of T follicular helper cell differentiation - PubMed (original) (raw)
Bcl6 and Blimp-1 are reciprocal and antagonistic regulators of T follicular helper cell differentiation
Robert J Johnston et al. Science. 2009.
Abstract
Effective B cell-mediated immunity and antibody responses often require help from CD4+ T cells. It is thought that a distinct CD4+ effector T cell subset, called T follicular helper cells (T(FH)), provides this help; however, the molecular requirements for T(FH) differentiation are unknown. We found that expression of the transcription factor Bcl6 in CD4+ T cells is both necessary and sufficient for in vivo T(FH) differentiation and T cell help to B cells in mice. In contrast, the transcription factor Blimp-1, an antagonist of Bcl6, inhibits T(FH) differentiation and help, thereby preventing B cell germinal center and antibody responses. These findings demonstrate that T(FH) cells are required for proper B cell responses in vivo and that Bcl6 and Blimp-1 play central but opposing roles in T(FH) differentiation.
Figures
Fig. 1
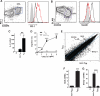
Bcl6 is a TFH-specific transcription factor. Naïve SMtg CD4+ T cells were transferred into B6 mice. In all panels, splenocytes were analyzed 8 days after infection with LCMV. (A and B) SMtg expression of CXCR5 and PD-1 (A) or SLAM (CD150) (B). SMtg+ (CD45.1+) CD4+ gated cells are shown. CXCR5high TFH cells are boxed in fluorescence-activated cell sorter (FACS) plots. Histogram overlays depict TFH cells (red) as well as naïve CD4+ T cells (gray) and CXCR5low non-TFH SMtg cells (black). Data are representative of more than 10 independent experiments. (C) IL-21 mRNA in SMtg CD4+ T cells, normalized to the β-actin mRNA level (×10–4). **P = 0.008. (D) In vitro chemotaxis toward CXCL13 (BLC) by ex vivo SMtg CD4+ T cells. Results are expressed as percentages of SLAMlow TFH SMtg (solid circles) and SLAMhigh non-TFH SMtg (open circles) that migrated in a transwell assay. ***P ≤ 0.001. 1 μg, P = 0.001; 2 μg, P = 0.0006; 4 μg, P = 0.0006. Data are representative of three independent experiments; n = 2 per group. (E) Scatterplot of the average signal of biological replicates of TFH versus non-TFH SMtg gene expression microarray data. Blue lines indicate changes in gene expression by a factor of 3; 386 gene probes exhibited a factor of >3.0 increase in TFH. Data from one of two independent experiments are shown; n = 2 per group. (F and G) Quantitative reverse transcription PCR of Bcl6 (F) and Blimp-1 (G) mRNA expression, normalized to β-actin (×10–4). ***P < 0.0001. Data are representative of four independent experiments; n = 2 per group. Error bars in all graphs are SEM.
Fig. 2
Bcl6 expression is sufficient for TFH differentiation in vivo. (A to E) Naïve SMtg CD4+ T cells were transduced with Bcl6-RV (Bcl6+) or left untransduced (control) and transferred into B6 mice subsequently infected with LCMV. (A) Gating of CD45.1+ untransduced SMtg (GFP–) and Bcl6-RV+ SMtg (GFP+) in the same host. CD4+ B220– gate is shown. (B) TFH (SLAMlow CXCR5high, boxed) and non-TFH (SLAMhigh CXCR5low) differentiation of untransduced SMtg (left) and Bcl6-RV+ SMtg (right). (C) Quantitation of SMtg TFH differentiation. Mice received Bcl6-RV+ SMtg and untransduced SMtg, or GFP-RV+ and untransduced SMtg. “–,” untransduced; “+,” transduced with indicated RV. ***P < 0.0001. Data are representative of three independent experiments; _n_ = 4 per group. (D and E) CXCR5 expression (D) and PD-1 expression (E) on naïve CD4+ T cells (gray), GFP-RV+ SMtg (black), and Bcl6-RV+ SMtg (red). Bar graphs show mean fluorescence intensity (MFI) of naïve CD4+ T cells, GFP-RV+ SMtg non-TFH, GFP-RV+ SMtg TFH, and Bcl6-RV+ SMtg. For PD-1 MFI, non-TFH versus TFH or Bcl6-RV+, _P_ < 0.05; TFH versus Bcl6-RV+, _P_ > 0.05. Data are representative of three independent experiments; n = 4 to 6 per group. (F and G) GFP-RV+ or Bcl6-RV+ SMtg cells were adoptively transferred separately into B cell–deficient mice (μMT) or HEL-specific BCR transgenic mice (MD4) on a μMT background. Host mice were subsequently infected with LCMV. Each group is a composite of three experiments; n = 2 (GFP-RV+ μMT), 6 (Bcl6-RV+ MD4), 6 (Bcl6-RV+ μMT), or 8 (GFP-RV+ MD4) per group. (F) Differentiation of SMtg CD4+ T cells in MD4 BCR transgenic mice. TFH cells (SLAMlow CXCR5high) are boxed. CD4+ B220– CD45.1+ GFP+ gate is shown. (G) Quantitation of SMtg TFH differentiation. GFP-RV+ versus Bcl6-RV+ in MD4, ***P < 0.0001. GFP-RV+ versus Bcl6-RV+ in μMT, ***P < 0.0001.
Fig. 3
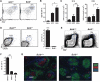
Bcl6 expression is necessary for inducing TFH B cell help in vivo. (A) Germinal center B cells (PNA+ Fas+, gated) in mice that received GFP-RV+ or Bcl6-RV+ SMtg CD4+ T cells and were subsequently infected with LCMV, analyzed at day 8 and gated on activated B cells (B220+ IgDlow). (B) Frequency of germinal center B cells of total splenocytes; n = 4 per group. Data are representative of three independent experiments. *P = 0.029. (C) GFP-RV+ or Bcl6-RV+ OT-II CD4+ T cells were transferred into B6 mice subsequently immunized with NP-Ova in alum. Control mice were immunized but received no OT-II cells. NP-Ova enzyme-linked immunosorbent assay (ELISA) was performed at day 15 and day 45; n = 6 per group. Data are representative of two independent experiments. Day 15 endpoint ELISA titers, **P = 0.008; day 45 endpoint ELISA titers, *P = 0.017.(D) Bcl6+/+ or _Bcl6_–/– OT-II CD4+ Tcells were transferred into congenically mismatched B6 mice subsequently immunized with Ova in alum. Splenocytes were analyzed 6 days after immunization; n = 4 per group. Data are representative of four independent experiments. OT-II+ CD44high gate is shown. Quantitation of OT-II TFH differentiation is also shown. ***P < 0.0001. (E to G) Bcl6+/+ or _Bcl6_–/– OT-II CD4+ T cells were cotransferred with B1-8 B cells into _Icos_–/– mice subsequently immunized with NP-Ova in alum; n = 2 per group. Data are representative of two independent experiments. (E) Germinal center B cells (PNA+ GL7+, boxed) 7 days after immunization. TCRβ– IgDlow gate is shown. (F) Quantitation of GC B cells as percent of spleen. **P = 0.0015. (G) Germinal center histology. Spleen sections were stained with IgD (green), PNA (red), and CD4 (blue).
Fig. 4
Blimp-1 and Bcl6 are antagonistic and reciprocal regulators of TFH differentiation. (A) Immunoblot of Bcl6 protein expression (and β-actin control) in transduced SMtg CD4+ T cells in vivo. (B) TFH (SLAMlow CXCR5high, boxed) differentiation of untransduced SMtg (left, “Control”) and Blimp1-RV+ SMtg (right, “Blimp-1+”) cells within a common host 8 days after LCMV infection. Gating is shown in fig. S10B. (C) Quantitation of SMtg TFH differentiation. “–,” untransduced; “+,” transduced with the indicated RV. ***P < 0.0001; n = 4 per group. Data are representative of two independent experiments. (D) SLAM and CXCR5 expression by naïve CD4+ T cells (gray), GFP-RV+ SMtg (black), and Blimp1-RV+ SMtg (red). (E and F) GFP-RV+ or Blimp1-RV+ OT-II CD4+ T cells were transferred into SAP-deficient mice subsequently immunized with NP-Ova in alum; n = 4 per group. Data are representative of three independent experiments. (E) Germinal center B cells (PNA+ Fas+, gated) in mice that received GFP-RV+ or Blimp1-RV+ OT-II CD4+ T cells. B220+ IgDlow gate is shown. Quantitation of germinal center B cells in the spleen is also shown. ***P < 0.0001. (F) NP-Ova IgG ELISA endpoint titers at day 10. *P = 0.016. (G) Purified naïve B6 and prdm1fl/fl CD45.2+ CD4+ T cells were transduced with SMtg-RV, with or without Cre-RV (Cre+ or Cre–), sorted, and transferred into CD45.1+ mice subsequently infected with LCMV. FACS plots depict TFH (CXCR5high SLAMlow, boxed) differentiation of control Cre+ SMtg+ B6 cells (left), Blimp-1–sufficient Cre– SMtg+ prdm1fl/fl cells (center), and Blimp-1–deficient Cre+ SMtg+ prdm1fl/fl cells (right). CD4+ CD45.1– CD44high 7AAD– gate is shown. Quantitation of TFH differentiation is also shown. Data are representative of two independent experiments. **P = 0.002, ***P = 0.0006; n = 4 to 5 per group.
Comment in
- Immunology. The yin and yang of follicular helper T cells.
Awasthi A, Kuchroo VK. Awasthi A, et al. Science. 2009 Aug 21;325(5943):953-5. doi: 10.1126/science.1178752. Science. 2009. PMID: 19696339 No abstract available.
Similar articles
- Bcl6 mediates the development of T follicular helper cells.
Nurieva RI, Chung Y, Martinez GJ, Yang XO, Tanaka S, Matskevitch TD, Wang YH, Dong C. Nurieva RI, et al. Science. 2009 Aug 21;325(5943):1001-5. doi: 10.1126/science.1176676. Epub 2009 Jul 23. Science. 2009. PMID: 19628815 Free PMC article. - STAT5 is a potent negative regulator of TFH cell differentiation.
Johnston RJ, Choi YS, Diamond JA, Yang JA, Crotty S. Johnston RJ, et al. J Exp Med. 2012 Feb 13;209(2):243-50. doi: 10.1084/jem.20111174. Epub 2012 Jan 23. J Exp Med. 2012. PMID: 22271576 Free PMC article. - In vivo regulation of Bcl6 and T follicular helper cell development.
Poholek AC, Hansen K, Hernandez SG, Eto D, Chandele A, Weinstein JS, Dong X, Odegard JM, Kaech SM, Dent AL, Crotty S, Craft J. Poholek AC, et al. J Immunol. 2010 Jul 1;185(1):313-26. doi: 10.4049/jimmunol.0904023. Epub 2010 Jun 2. J Immunol. 2010. PMID: 20519643 Free PMC article. - IL-21 and T follicular helper cells.
Spolski R, Leonard WJ. Spolski R, et al. Int Immunol. 2010 Jan;22(1):7-12. doi: 10.1093/intimm/dxp112. Epub 2009 Nov 23. Int Immunol. 2010. PMID: 19933709 Free PMC article. Review. - Follicular helper CD4 T cells (TFH).
Crotty S. Crotty S. Annu Rev Immunol. 2011;29:621-63. doi: 10.1146/annurev-immunol-031210-101400. Annu Rev Immunol. 2011. PMID: 21314428 Review.
Cited by
- Multistability and predominant hybrid phenotypes in a four node mutually repressive network of Th1/Th2/Th17/Treg differentiation.
Duddu AS, Andreas E, Bv H, Grover K, Singh VR, Hari K, Jhunjhunwala S, Cummins B, Gedeon T, Jolly MK. Duddu AS, et al. NPJ Syst Biol Appl. 2024 Oct 24;10(1):123. doi: 10.1038/s41540-024-00433-6. NPJ Syst Biol Appl. 2024. PMID: 39448615 Free PMC article. - Interleukin 9 mediates T follicular helper cell activation to promote antibody responses.
Sato T, Ikegami I, Yanagi M, Ohyu T, Sugaya T, Shirato S, Tanemoto M, Kamiya S, Kikuchi K, Kamada Y, Nakata T, Kamekura R, Sato A, Takano KI, Miyajima M, Watanabe A, Ichimiya S. Sato T, et al. Front Immunol. 2024 Sep 30;15:1441407. doi: 10.3389/fimmu.2024.1441407. eCollection 2024. Front Immunol. 2024. PMID: 39403384 Free PMC article. - Role of circulating T follicular helper subsets following Ty21a immunization and oral challenge with wild type S. Typhi in humans.
Booth JS, Rapaka RR, McArthur MA, Fresnay S, Darton TC, Blohmke CJ, Jones C, Waddington CS, Levine MM, Pollard AJ, Sztein MB. Booth JS, et al. Front Immunol. 2024 Sep 12;15:1384642. doi: 10.3389/fimmu.2024.1384642. eCollection 2024. Front Immunol. 2024. PMID: 39328410 Free PMC article. - Role of cellular effectors in the induction and maintenance of IgA responses leading to protective immunity against enteric bacterial pathogens.
Carreto-Binaghi LE, Sztein MB, Booth JS. Carreto-Binaghi LE, et al. Front Immunol. 2024 Sep 11;15:1446072. doi: 10.3389/fimmu.2024.1446072. eCollection 2024. Front Immunol. 2024. PMID: 39324143 Free PMC article. Review. - Clinical significance of T helper cell subsets in the peripheral blood and bone marrow of patients with multiple myeloma.
Zhang L, Zhong H, Fan J, Mao J, Li Y. Zhang L, et al. Front Immunol. 2024 Sep 11;15:1445530. doi: 10.3389/fimmu.2024.1445530. eCollection 2024. Front Immunol. 2024. PMID: 39324138 Free PMC article.
References
- King C, Tangye S, Mackay C. Annu. Rev. Immunol. 2008;26:741. - PubMed
- Vinuesa CG, et al. Nature. 2005;435:452. - PubMed
- Chtanova T, et al. J. Immunol. 2004;173:68. - PubMed
- Rasheed AU, Rahn HP, Sallusto F, Lipp M, Müller G. Eur. J. Immunol. 2006;36:1892. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R37 AR040072/AR/NIAMS NIH HHS/United States
- R01 AI063107-01A1/AI/NIAID NIH HHS/United States
- P30 AR053495/AR/NIAMS NIH HHS/United States
- AR40072/AR/NIAMS NIH HHS/United States
- R01 072543/PHS HHS/United States
- R01 AI063107/AI/NIAID NIH HHS/United States
- R01 AI072543-01A1/AI/NIAID NIH HHS/United States
- AR44076/AR/NIAMS NIH HHS/United States
- R01 AI072543/AI/NIAID NIH HHS/United States
- R01 063107/PHS HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous