Reversal of RNA dominance by displacement of protein sequestered on triplet repeat RNA - PubMed (original) (raw)
Reversal of RNA dominance by displacement of protein sequestered on triplet repeat RNA
Thurman M Wheeler et al. Science. 2009.
Abstract
Genomic expansions of simple tandem repeats can give rise to toxic RNAs that contain expanded repeats. In myotonic dystrophy, the expression of expanded CUG repeats (CUGexp) causes abnormal regulation of alternative splicing and neuromuscular dysfunction. We used a transgenic mouse model to show that derangements of myotonic dystrophy are reversed by a morpholino antisense oligonucleotide, CAG25, that binds to CUGexp RNA and blocks its interaction with muscleblind-like 1 (MBNL1), a CUGexp-binding protein. CAG25 disperses nuclear foci of CUGexp RNA and reduces the overall burden of this toxic RNA. As MBNL1 is released from sequestration, the defect of alternative splicing regulation is corrected, thereby restoring ion channel function. These findings suggest an alternative use of antisense methods, to inhibit deleterious interactions of proteins with pathogenic RNAs.
Figures
Fig. 1
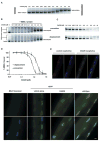
CAG25 inhibits formation of CUGexp-MBNL1 complexes. (A) Gel shift assay demonstrates that addition of CAG25 (indicated concentration) to labeled (CUG)109 (2 nM) results in slower migration of (CUG)109-CAG25 heteroduplex. (B) CAG25 prevents CUGexp-MBNL1 complex formation (upper panel) and displaces MBNL1 protein from pre-formed complex (lower panel). Lane “C” shows migration of labeled (CUG)109 hairpin. Addition of excess MBNL1 produces complexes of variable size that migrate as a broad smear (lane “0.0”). Addition of CAG25 at increasing concentration reconstitutes (CUG)109-CAG25 heteroduplex as the dominant band (27). (C) Microtiter plate/gel assay confirms that CAG25 prevents the formation of (CUG)109-MBNL1 complex (upper panel) and displaces MBNL1 from pre-formed complexes (lower panel). Bands indicate the amount of recombinant MBNL1 protein that remains in ribonucleoprotein complex at the indicated concentration of CAG25 (27). (D) The % MBNL1 bound to CUGexp is expressed as the mean ± SD of protein retained on plate. IC50 for “prevention” is 462 ± 31 nM and for “displacement” is 1032 ± 117 nM. (E) Fluorescence in situ hybridization of single flexor digitorum brevis (FDB) muscle fibers from _HSA_LR mice. Probe (red) binds to _HSA_LR transcripts upstream from CUG repeat, nuclei are blue. CAG25, but not control morpholino, causes dispersal of RNA foci. (F) MBNL1 (immunofluorescence, green) shifts from punctate to diffuse nuclear distribution after injection of FDB with CAG25. Scale bars = 5 μm.
Fig. 2
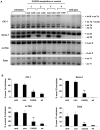
Reversal of misregulated alternative splicing by CAG25. (A) RT-PCR analysis of alternative splicing for chloride channel-1 (ClC-1), Serca-1, m-Titin, and Zasp, 3 weeks after a single injection of CAG25 in tibialis anterior (TA) muscle of _HSA_LR transgenic mice. The contralateral (con) TA was injected with vehicle (saline, mice 1 and 2) or morpholino with inverted sequence (GAC25, mice 3 and 4). Splice products from untreated _HSA_LR transgenic and wild-type TA muscle (n = 3 mice) are shown. Int 6 = intron 6 retention. (B) Quantification of results in A, expressed as the percentage of splice products that include or exclude the indicated exon. *P = 0.003 and ** P < 0.001 for CAG25 vs contralateral (_t_-test). “untr” = untreated _HSA_LR.
Fig. 3
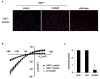
Chloride channel 1 protein expression and function are rescued by CAG25. (A) Immunofluorescence for ClC-1 protein expression in sections from _HSA_LR TA muscle, bar = 20 μm. (B) ClC-1 current density in FDB fibers isolated from 15-day-old _HSA_LR mice injected with CAG25 or control morpholino. Fibers from wild-type mice injected with CAG25 or control morpholino serve as controls (see also Fig. S6). (C) Electromyographic myotonia analysis 3 weeks after injection of CAG25 into TA muscle. The contralateral side was injected with vehicle (saline) alone or control morpholino (27). n = 11 mice examined; *P < 0.0001 for CAG25 vs. saline (_t_-test).
Fig. 4
CAG25 increases the cytoplasmic level and translation of CUGexp-containing mRNA, despite reducing the overall level of CUGexp RNA. (A) RT-PCR assay of transgene mRNA in the cytoplasm. Human (transgene) and mouse (endogenous) skeletal actin transcripts were co-amplified by the same primers, species origin was revealed by AluI cleavage (27). “h”, human muscle; “wt”, wild-type mouse. Lower panel shows depletion of nuclear RNA from cytoplasmic fraction (“c”) relative to nuclear pellet (“n”), analyzed by RT-PCR for the 5′ external transcribed spacer (5′ ETS, nuclear-retained) of ribosomal RNA. Numbers refer to different mice treated with CAG25. (B) CAG25 increases luciferase activity in LLC9/Rosa-CreER bitransgenic mice. Upper panel shows LLC9 transgene for conditional expression of CUGexp RNA. Lower panels show in vivo bioluminescence imaging (BLI) of different bitransgenic mice (27). For quantification, luciferase activity (indicated in yellow-orange) in CAG25-injected muscle was normalized to the contralateral side injected with saline or control morpholino (n = 7 mice). (C) Northern blot of total cellular RNA shows decreased _HSA_LR mRNA in muscle injected with CAG25, compared to contralateral muscle injected with vehicle (saline). Mouse actin is loading control. (D) CUGexp-CAG25 heteroduplex is not cleaved by RNase H. (CUG)109 RNA was incubated with CAG25 morpholino or DNA oligonucleotide of identical sequence (CAG25-DNA). The heteroduplex was incubated with the indicated concentration of RNase H, then separated on polyacrylamide gels. Lane “c”, control with (CUG)109 alone. (E) (CUG)109 RNA was incubated with CAG25 morpholino or CAG25-DNA to form heteroduplex as in D, to which HeLa cell extract was added for the indicated time.
Comment in
- Molecular biology. Neutralizing toxic RNA.
Cooper TA. Cooper TA. Science. 2009 Jul 17;325(5938):272-3. doi: 10.1126/science.1177452. Science. 2009. PMID: 19608901 No abstract available.
Similar articles
- Molecular biology. Neutralizing toxic RNA.
Cooper TA. Cooper TA. Science. 2009 Jul 17;325(5938):272-3. doi: 10.1126/science.1177452. Science. 2009. PMID: 19608901 No abstract available. - Systemic delivery of a Peptide-linked morpholino oligonucleotide neutralizes mutant RNA toxicity in a mouse model of myotonic dystrophy.
Leger AJ, Mosquea LM, Clayton NP, Wu IH, Weeden T, Nelson CA, Phillips L, Roberts E, Piepenhagen PA, Cheng SH, Wentworth BM. Leger AJ, et al. Nucleic Acid Ther. 2013 Apr;23(2):109-17. doi: 10.1089/nat.2012.0404. Epub 2013 Jan 11. Nucleic Acid Ther. 2013. PMID: 23308382 - RNA interference targeting CUG repeats in a mouse model of myotonic dystrophy.
Sobczak K, Wheeler TM, Wang W, Thornton CA. Sobczak K, et al. Mol Ther. 2013 Feb;21(2):380-7. doi: 10.1038/mt.2012.222. Epub 2012 Nov 27. Mol Ther. 2013. PMID: 23183533 Free PMC article. - Gain of RNA function in pathological cases: Focus on myotonic dystrophy.
Klein AF, Gasnier E, Furling D. Klein AF, et al. Biochimie. 2011 Nov;93(11):2006-12. doi: 10.1016/j.biochi.2011.06.028. Epub 2011 Jul 13. Biochimie. 2011. PMID: 21763392 Review. - Pathogenic mechanisms of myotonic dystrophy.
Lee JE, Cooper TA. Lee JE, et al. Biochem Soc Trans. 2009 Dec;37(Pt 6):1281-6. doi: 10.1042/BST0371281. Biochem Soc Trans. 2009. PMID: 19909263 Free PMC article. Review.
Cited by
- A Potent Inhibitor of Protein Sequestration by Expanded Triplet (CUG) Repeats that Shows Phenotypic Improvements in a Drosophila Model of Myotonic Dystrophy.
Luu LM, Nguyen L, Peng S, Lee J, Lee HY, Wong CH, Hergenrother PJ, Chan HY, Zimmerman SC. Luu LM, et al. ChemMedChem. 2016 Jul 5;11(13):1428-35. doi: 10.1002/cmdc.201600081. Epub 2016 Jun 1. ChemMedChem. 2016. PMID: 27245480 Free PMC article. - Generation of neural cells from DM1 induced pluripotent stem cells as cellular model for the study of central nervous system neuropathogenesis.
Xia G, Santostefano KE, Goodwin M, Liu J, Subramony SH, Swanson MS, Terada N, Ashizawa T. Xia G, et al. Cell Reprogram. 2013 Apr;15(2):166-77. doi: 10.1089/cell.2012.0086. Cell Reprogram. 2013. PMID: 23550732 Free PMC article. Clinical Trial. - Ultrasound-enhanced delivery of morpholino with Bubble liposomes ameliorates the myotonia of myotonic dystrophy model mice.
Koebis M, Kiyatake T, Yamaura H, Nagano K, Higashihara M, Sonoo M, Hayashi Y, Negishi Y, Endo-Takahashi Y, Yanagihara D, Matsuda R, Takahashi MP, Nishino I, Ishiura S. Koebis M, et al. Sci Rep. 2013;3:2242. doi: 10.1038/srep02242. Sci Rep. 2013. PMID: 23873129 Free PMC article. - Identification of Plant-derived Alkaloids with Therapeutic Potential for Myotonic Dystrophy Type I.
Herrendorff R, Faleschini MT, Stiefvater A, Erne B, Wiktorowicz T, Kern F, Hamburger M, Potterat O, Kinter J, Sinnreich M. Herrendorff R, et al. J Biol Chem. 2016 Aug 12;291(33):17165-77. doi: 10.1074/jbc.M115.710616. Epub 2016 Jun 13. J Biol Chem. 2016. PMID: 27298317 Free PMC article. - RNA-mediated neurodegeneration in repeat expansion disorders.
Todd PK, Paulson HL. Todd PK, et al. Ann Neurol. 2010 Mar;67(3):291-300. doi: 10.1002/ana.21948. Ann Neurol. 2010. PMID: 20373340 Free PMC article. Review.
References
- Mankodi A, et al. Science. 2000;289:1769. - PubMed
- Liquori CL, et al. Science. 2001;293:864. - PubMed
- Jin P, et al. Neuron. 2003;39:739. - PubMed
- Moseley ML, et al. Nat Genet. 2006;38:758. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- NIDCR-T32DE07202/DE/NIDCR NIH HHS/United States
- K08 NS064293/NS/NINDS NIH HHS/United States
- T32 DE007202/DE/NIDCR NIH HHS/United States
- R01 AR049077/AR/NIAMS NIH HHS/United States
- R01 AR046806/AR/NIAMS NIH HHS/United States
- AR/NS48143/AR/NIAMS NIH HHS/United States
- AR046806/AR/NIAMS NIH HHS/United States
- K24 AR048143/AR/NIAMS NIH HHS/United States
- U54 NS048843/NS/NINDS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials