Selective anchoring of DNA methyltransferases 3A and 3B to nucleosomes containing methylated DNA - PubMed (original) (raw)
Selective anchoring of DNA methyltransferases 3A and 3B to nucleosomes containing methylated DNA
Shinwu Jeong et al. Mol Cell Biol. 2009 Oct.
Abstract
Proper DNA methylation patterns are essential for mammalian development and differentiation. DNA methyltransferases (DNMTs) primarily establish and maintain global DNA methylation patterns; however, the molecular mechanisms for the generation and inheritance of methylation patterns are still poorly understood. We used sucrose density gradients of nucleosomes prepared by partial and maximum micrococcal nuclease digestion, coupled with Western blot analysis to probe for the interactions between DNMTs and native nucleosomes. This method allows for analysis of the in vivo interactions between the chromatin modification enzymes and their actual nucleosomal substrates in the native state. We show that little free DNA methyltransferase 3A and 3B (DNMT3A/3B) exist in the nucleus and that almost all of the cellular contents of DNMT3A/3B, but not DNMT1, are strongly anchored to a subset of nucleosomes. This binding of DNMT3A/3B does not require the presence of other well-known chromatin-modifying enzymes or proteins, such as proliferating cell nuclear antigen, heterochromatin protein 1, methyl-CpG binding protein 2, Enhancer of Zeste homolog 2, histone deacetylase 1, and UHRF1, but it does require an intact nucleosomal structure. We also show that nucleosomes containing methylated SINE and LINE elements and CpG islands are the main sites of DNMT3A/3B binding. These data suggest that inheritance of DNA methylation requires cues from the chromatin component in addition to hemimethylation.
Figures
FIG. 1.
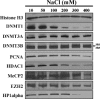
DNMT3A/3B, but not DNMT1, are strongly associated with chromatin in nuclei. Nuclei purified from 5 × 106 HCT116 cells were incubated in nondenaturing extraction buffers containing 10 to 400 mM NaCl in the presence of protease inhibitors for 5 min. The nuclei were purified by centrifugation, and the proteins remaining within the nuclei were resolved by SDS-polyacrylamide gel electrophoresis and probed by Western blotting with various antibodies. The positions of two DNMT3B isoforms (DNMT3B2 and DNMT3B3) are indicated to the right of the blots.
FIG. 2.
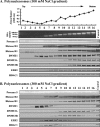
DNMT3A/3B are bound to polynucleosomes at high ionic strength. Nucleosomes released from nuclei partially digested with MNase at low ionic strength (20 mM) were resolved by ultracentrifugation on a sucrose density gradient (5% to 25%) containing 100 mM NaCl (A) and 300 mM NaCl (B). Gradients were fractionated into 16 aliquots numbered 1 to 16 starting from the top of the centrifuge tube. The absorbance of each fraction was read at 260 nm. DNA purified from each fraction was resolved by agarose gel electrophoresis and stained with EtBr. To probe the distribution of proteins in each fraction, Western blotting was performed with various antibodies. Ponceau S staining shows core histones transferred onto the membrane from the SDS-polyacrylamide gel. The control lanes on the gels were loaded with unfractionated nuclear extract loaded on the gels to monitor the quality of immunostaining of the membranes.
FIG. 3.
DNMT3A/3B binding requires an intact nucleosomal structure. Mononucleosomes released by extensive digestion with MNase were resolved on a sucrose density gradient (5% to 25%) containing 300 mM NaCl. Mononucleosomes were incubated in the absence (A) or presence (B) of 300 μg/ml EtBr for 10 min at room temperature before they were loaded onto the gradients. The gradients were fractionated and analyzed as described in the legend to Fig. 2.
FIG. 4.
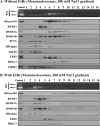
DNMT1 forms a stable complex in the linker DNA region with 5-aza-2′-deoxycytidine. HCT116 cells were cultured in the presence of 1 μM 5-aza-CdR for 24 h, followed by nucleus preparation. Polynucleosomes released from the nuclei digested with MNase were resolved through a sucrose density gradient (5% to 25%) containing 300 mM NaCl. DNA preparation and Western blotting were performed as described in the legend to Fig. 2.
FIG. 5.
The N-terminal region of DNMT3B is necessary for strong nucleosomal binding. (A) Map of DNMT3A/3B isoforms showing the PWWP and PHD-like domains located in the N-terminal regions and the catalytic methylase domains in the C-terminal region. (B) Mononucleosomes from 293T cells transfected with expression vectors of various Myc-DNMT3A/3B deletion proteins were subjected to sucrose gradients containing 300 mM NaCl. Endogenous and exogenous enzymes on the gradient were detected by Western blot analysis using specific antibodies against endogenous protein and anti-Myc antibody. Gradients were analyzed as described in the legend to Fig. 2.
FIG. 6.
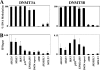
DNMT3A/3B are enriched in highly DNA methylated CpG islands. (A) The levels of DNA methylation in nine different CpG islands in 293T cells transfected with myc-DNMT3A1 or DNMT3B1 constructs were determined by the methylation-sensitive single-nucleotide primer extension assay. (B) ChIP assays were performed with anti-DNMT3A antibodies for the cells with myc-DNMT3A1 or anti-Myc antibody for the cells with myc-DNMT3B1. Values are the averages of at least triplicate determinations with standard errors (error bars) indicated.
FIG. 7.
DNMT3A/3B preferentially associate with methylated repetitive DNA elements and CpG islands, but not with nuclear RNA. (A) Western blot analysis was performed to analyze the proteins immunoprecipitated using DNMT3A, DNMT3B, and control CD8 antibodies. DNMT3A/3B antibodies specifically pulled down DNMT3A/3B proteins. (B) Nucleic acids (DNA and/or RNA, if any) pulled down by DNMT3A and CD8 antibodies from pooled polynucleosomal fractions from a partially digested sucrose gradient were 32P end labeled, treated with DNase or RNase or not treated, and then analyzed on a denaturing polyacrylamide/urea electrophoresis gel. The positions of trinucleosomes (Tri-Nuc), dinucleosomes (Di-Nuc), and mononucleosomes (Mono-Nuc) and molecular size markers are indicated to the left of the gel. (C) Distribution of different sequence classes in samples obtained by DNMT3A/3B IP. Sequence analysis of DNA fragments isolated by genome-wide amplification (GWA) of DNA precipitated by a mixture of DNMT3 antibodies shows preferential association of DNMT3A/3B with repetitive elements, such as SINES, LINES, etc. (D) Selective amplification of the DNA in the IP material through AP-PCR using GC-rich primers (ChAP assay [ChIP assay coupled with an arbitrarily primed PCR]) also shows the presence of repeats and single-copy CGIs. (E) Native ChIP assays were performed with DNMT3A/3B-specific antibodies followed by DNA purification from input, unbound (UB), and immunoprecipitated (IP) samples. The DNA samples were then digested with buffer alone (U), MspI (M), and HpaII (H) and subsequently used for an AP-PCR. The PCR products were resolved on a sequencing polyacrylamide gel, and part of the gel is shown as an example. The two strands of the same fragment sometimes resolved as separate bands on the gel due to the slight difference in their molecular weights. Fourteen informative bands were then extracted from the gel and sequenced. The numbers to the right of the gel denote the corresponding fragment number in panel F. (F) Properties of DNA fragments containing HpaII sites that were isolated from the sequencing polyacrylamide gel.
Similar articles
- Nucleosomes containing methylated DNA stabilize DNA methyltransferases 3A/3B and ensure faithful epigenetic inheritance.
Sharma S, De Carvalho DD, Jeong S, Jones PA, Liang G. Sharma S, et al. PLoS Genet. 2011 Feb 3;7(2):e1001286. doi: 10.1371/journal.pgen.1001286. PLoS Genet. 2011. PMID: 21304883 Free PMC article. - DNA methyltransferase 3b preferentially associates with condensed chromatin.
Kashiwagi K, Nimura K, Ura K, Kaneda Y. Kashiwagi K, et al. Nucleic Acids Res. 2011 Feb;39(3):874-88. doi: 10.1093/nar/gkq870. Epub 2010 Oct 4. Nucleic Acids Res. 2011. PMID: 20923784 Free PMC article. - Cooperativity between DNA methyltransferases in the maintenance methylation of repetitive elements.
Liang G, Chan MF, Tomigahara Y, Tsai YC, Gonzales FA, Li E, Laird PW, Jones PA. Liang G, et al. Mol Cell Biol. 2002 Jan;22(2):480-91. doi: 10.1128/MCB.22.2.480-491.2002. Mol Cell Biol. 2002. PMID: 11756544 Free PMC article. - The Roles of Human DNA Methyltransferases and Their Isoforms in Shaping the Epigenome.
Gujar H, Weisenberger DJ, Liang G. Gujar H, et al. Genes (Basel). 2019 Feb 23;10(2):172. doi: 10.3390/genes10020172. Genes (Basel). 2019. PMID: 30813436 Free PMC article. Review. - Mammalian DNA methyltransferases: new discoveries and open questions.
Gowher H, Jeltsch A. Gowher H, et al. Biochem Soc Trans. 2018 Oct 19;46(5):1191-1202. doi: 10.1042/BST20170574. Epub 2018 Aug 28. Biochem Soc Trans. 2018. PMID: 30154093 Free PMC article. Review.
Cited by
- Kinetics of DNA methylation inheritance by the Dnmt1-including complexes during the cell cycle.
Hervouet E, Nadaradjane A, Gueguen M, Vallette FM, Cartron PF. Hervouet E, et al. Cell Div. 2012 Feb 20;7:5. doi: 10.1186/1747-1028-7-5. Cell Div. 2012. PMID: 22348533 Free PMC article. - Pluripotency transcription factor Oct4 mediates stepwise nucleosome demethylation and depletion.
Shakya A, Callister C, Goren A, Yosef N, Garg N, Khoddami V, Nix D, Regev A, Tantin D. Shakya A, et al. Mol Cell Biol. 2015 Mar;35(6):1014-25. doi: 10.1128/MCB.01105-14. Epub 2015 Jan 12. Mol Cell Biol. 2015. PMID: 25582194 Free PMC article. - Viral Manipulation of the Host Epigenome as a Driver of Virus-Induced Oncogenesis.
Soliman SHA, Orlacchio A, Verginelli F. Soliman SHA, et al. Microorganisms. 2021 May 30;9(6):1179. doi: 10.3390/microorganisms9061179. Microorganisms. 2021. PMID: 34070716 Free PMC article. Review. - Enzymology of Mammalian DNA Methyltransferases.
Jurkowska RZ, Jeltsch A. Jurkowska RZ, et al. Adv Exp Med Biol. 2022;1389:69-110. doi: 10.1007/978-3-031-11454-0_4. Adv Exp Med Biol. 2022. PMID: 36350507 - Rethinking how DNA methylation patterns are maintained.
Jones PA, Liang G. Jones PA, et al. Nat Rev Genet. 2009 Nov;10(11):805-11. doi: 10.1038/nrg2651. Epub 2009 Sep 30. Nat Rev Genet. 2009. PMID: 19789556 Free PMC article. Review.
References
- Aapola, U., K. Kawasaki, H. S. Scott, J. Ollila, M. Vihinen, M. Heino, A. Shintani, K. Kawasaki, S. Minoshima, K. Krohn, S. E. Antonarakis, N. Shimizu, J. Kudoh, and P. Peterson. 2000. Isolation and initial characterization of a novel zinc finger gene, DNMT3L, on 21q22.3, related to the cytosine-5-methyltransferase 3 gene family. Genomics 65:293-298. - PubMed
- Arita, K., M. Ariyoshi, H. Tochio, Y. Nakamura, and M. Shirakawa. 2008. Recognition of hemi-methylated DNA by the SRA protein UHRF1 by a base-flipping mechanism. Nature 455:818-821. - PubMed
- Avvakumov, G. V., J. R. Walker, S. Xue, Y. Li, S. Duan, C. Bronner, C. H. Arrowsmith, and S. Dhe-Paganon. 2008. Structural basis for recognition of hemi-methylated DNA by the SRA domain of human UHRF1. Nature 455:822-825. - PubMed
- Bachman, K. E., M. R. Rountree, and S. B. Baylin. 2001. Dnmt3a and Dnmt3b are transcriptional repressors that exhibit unique localization properties to heterochromatin. J. Biol. Chem. 276:32282-32287. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials