Developmental roles of 21 Drosophila transcription factors are determined by quantitative differences in binding to an overlapping set of thousands of genomic regions - PubMed (original) (raw)
doi: 10.1186/gb-2009-10-7-r80. Epub 2009 Jul 23.
Xiao-Yong Li, Jingyi Li, James B Brown, Hou Cheng Chu, Lucy Zeng, Brandi P Grondona, Aaron Hechmer, Lisa Simirenko, Soile V E Keränen, David W Knowles, Mark Stapleton, Peter Bickel, Mark D Biggin, Michael B Eisen
Affiliations
- PMID: 19627575
- PMCID: PMC2728534
- DOI: 10.1186/gb-2009-10-7-r80
Developmental roles of 21 Drosophila transcription factors are determined by quantitative differences in binding to an overlapping set of thousands of genomic regions
Stewart MacArthur et al. Genome Biol. 2009.
Abstract
Background: We previously established that six sequence-specific transcription factors that initiate anterior/posterior patterning in Drosophila bind to overlapping sets of thousands of genomic regions in blastoderm embryos. While regions bound at high levels include known and probable functional targets, more poorly bound regions are preferentially associated with housekeeping genes and/or genes not transcribed in the blastoderm, and are frequently found in protein coding sequences or in less conserved non-coding DNA, suggesting that many are likely non-functional.
Results: Here we show that an additional 15 transcription factors that regulate other aspects of embryo patterning show a similar quantitative continuum of function and binding to thousands of genomic regions in vivo. Collectively, the 21 regulators show a surprisingly high overlap in the regions they bind given that they belong to 11 DNA binding domain families, specify distinct developmental fates, and can act via different cis-regulatory modules. We demonstrate, however, that quantitative differences in relative levels of binding to shared targets correlate with the known biological and transcriptional regulatory specificities of these factors.
Conclusions: It is likely that the overlap in binding of biochemically and functionally unrelated transcription factors arises from the high concentrations of these proteins in nuclei, which, coupled with their broad DNA binding specificities, directs them to regions of open chromatin. We suggest that most animal transcription factors will be found to show a similar broad overlapping pattern of binding in vivo, with specificity achieved by modulating the amount, rather than the identity, of bound factor.
Figures
Figure 1
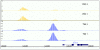
Similar patterns of in vivo DNA binding are detected by antibodies recognizing distinct epitopes on the same factor. The 675-bp window scores for ChIP/chip experiments across the rhomboid (rho) gene locus. Data are shown for pairs of antibodies against non-contigous portions of PRD and TWI proteins (Table 2). Nucleotide coordinates in the genome are given in base-pairs.
Figure 2
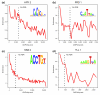
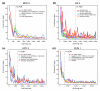
Recognition sequence enrichment correlates with ChIP/chip rank. Fold enrichment of matches to a position weight matrix (PWM) in the 500-bp windows around ChIP/chip peaks (± 250 bp), in non-overlapping cohorts of 200-peaks down the ChIP-chip rank list to the 25% FDR cutoff. Matches to the PWM below a _P_-value of ≤ 0.001 were scored. The PWMs used are shown as sequence logo representations [67]. The most highly bound peaks are to the left along the x-axis and the location of the 1% FDR threhold is indicated by a black, vertical dotted line. Shown are plots for the (a) HRY 2, (b) PRD 1, (c) SNA 2 and (d) TLL 1 antibodies.
Figure 3
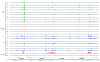
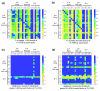
Broad, overlapping patterns of binding of transcription factors to the genome in blastoderm embryos. Data are shown for eight early A-P factors (green), six pair rule A-P factors (yellow), seven D-V factors (blue), and two general transcription factors (red). The 675-bp ChIP/chip window scores are plotted for regions bound above the 1% FDR threshold in a 500-kb portion of the genome. The locations of major RNA transcripts are shown below in grey for both DNA strands. The genome coordinates are given in base-pairs. For those factors for which ChIP/chip data are available for more than one antibody, data are shown for the antibody that gave the most bound regions above the 1% FDR threshold using the symmetric null test.
Figure 4
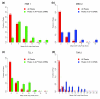
Known CRMs tend to be among the regions more highly bound in vivo. The 1% FDR bound regions for (a) HKB 1, (b) MED 2, (c) TLL 1 and (d) TWI were each divided into cohorts based on peak window score (x-axis). The fraction of all bound regions in each cohort (red bars) are shown (y-axis). In (a, c), the fraction of bound regions in each cohort in which the peak 500-bp window overlaps a CRM known to be regulated by at least some A-P early factors is shown (green bars). In (b, d), the fraction of bound regions that overlap a CRM known to be regulated by at least some D-V factors are shown (blue bars). The number of bound regions in each cohort is given above the bars.
Figure 5
Genes that control development are enriched in highly bound regions. The five most enriched Gene Ontology terms [68] in the 1% FDR bound regions for each factor were identified (enrichment measured by a hyper geometric test). The significance of the enrichment (-log(_P_-value)) of these five terms in non-overlapping cohorts of 200 peaks are shown down to the rank list as far as the 25% FDR cutoff. The most highly bound regions are to the left along the x-axis and the location of 1% FDR threshold is indicated by a black, vertical dotted line. Shown are the results for the (a) BCD 2, (b) DA 2, (c) HRY 2, and (d) RUN 1 antibodies. Dev., development; periph., peripheral; RNA pol, RNA polymerase; txn, transcription.
Figure 6
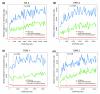
Highly bound regions are preferentially associated with genes transcribed and patterned in the blastoderm. Shown are the median distance of non-overlapping 200-peak cohorts to the closest gene belonging to each of three categories of gene: all genes (from genome release 4.3, March 2006; red lines); genes with known patterned expression (hand annotated based on Berkeley Drosophila Genome Project in situ images [23]; blue lines); and transcribed genes (defined by our RNA polymerase II (pol II) ChIP/chip binding [11]; green lines). Data are plotted down the ChIP/chip rank list to the 25% FDR threshold. The most highly bound regions are to the left along the x-axis and the location of 1% FDR threshold is indicated by a black, vertical dotted line. Shown are the results for the (a) DA 2, (b) HRY 2, (c) RUN 1, and (d) SNA 2 antibodies.
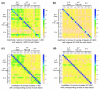
Figure 7
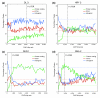
For some factors, poorly bound regions are preferentially found in protein coding sequences. The percentage of ChIP/chip peaks are plotted in non-overlapping cohorts of 200 peaks that are in protein coding (red), intronic (blue), and intergenic (green) sequences. Results are shown for cohorts down the rank lists to the 25% FDR cutoff. The percentages for each class of genomic feature are indicated as horizontal dotted lines in corresponding colors to the solid data lines. The most highly bound regions are to the left along the x-axis and the location of 1% FDR threshold is indicated by a black, vertical dotted line. Shown are the results for the (a) DL 3, (b) HRY 2, (c) RUN 1, and (d) SNA 2 antibodies.
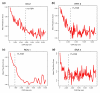
Figure 8
Highly bound regions are preferentially conserved. Mean PhastCons scores in the 500-bp windows (± 250 bp) around peaks, in non-overlapping cohorts of 200 peaks down the rank list towards the 25% FDR cutoff. The most highly bound peaks are to the left along the x-axis and the location of 1% FDR threshold is indicated by a black, vertical dotted line. Shown are the results for the (a) DA 2, (b) HRY 2, (c) RUN 1, and (d) SNA 2 antibodies.
Figure 9
Heat maps showing high overlap in binding among the blastoderm factors. (a, b) Each row shows the percentage of a cohort of 300 single nucleotide position peaks for a factor that are overlapped by 1% FDR regions bound by each of the other factors in turn. (c, d) Each row shows the Genome Structure Correction z scores for the likelihood that the overlap plotted in (a, b) occurs by chance given the proportion of the genome bound by each factor. (a, c) Results for the most highly bound 300 peaks (1-300). (b, d) Results for the second most highly bound cohort of 300 peaks (301-600). Note that the 1% FDR threshold does not lie within ranks 1 to 600 for 17 of the 21 factors shown, and, thus, for these proteins the bulk of the differences observed between the 1-300 and the 301-600 cohorts are not attributable to false positives.
Figure 10
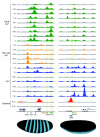
In vivo DNA binding of 21 sequence-specific and 2 general transcription factors to the even skipped (eve) and snail (sna) loci. ChIP/chip scores are plotted for 675-bp windows associated with all oligonucleotides on the array in the portions of the genome shown. In those regions bound above the 1% FDR threshold, the plots are colored green (Early A-P factors), yellow (Pair rule A-P factors), blue (D-V factors) or red (General factors). The locations of major RNA transcripts are shown below (blue) for both DNA strands together with the locations of CRMs active in blastoderm embryos (green) and later stages of development (salmon). Nucleotide coordinates in the genome are given in base-pairs. At the bottom is show the mRNA expression patterns of eve and sna in mid-stage 5 blastoderm embryos from the BDTNP's VirtualEmbryo using PointCloudXplore [12,69]. A more detailed plot comparing ChIP scores for both factor and negative control immunoprecipitations is shown in Additional data file 14, including data for all antibodies shown in Table 2.
Figure 11
The relative levels of eve and sna mRNA expression in mid-stage 5 blastoderm embryos at cellular resolution. Shown is a display from PointCloudXplore of a two-dimensional cylindrical projection of a VirtualEmbryo (D_mel_wt__atlas_r2.vpc) [12,69,70], where the level of mRNA expression is shown by height above a two-dimensional projection of the embryo surface. eve mRNA expression is shown in red and sna in green. The eve data are the average from images of 368 embryos and the sna data from 12 embryos.
Figure 12
Heat maps showing the binding of blastoderm transcription factors to validated A-P early and D-V CRMs. (a) Each row shows if a factor is detected binding or not to each CRM, where binding is defined as a 1% FDR region that overlaps the CRM by 500 bp or more. (b) Each row shows the ChIP/chip intensity of the highest 675-bp window for a factor on each of the 44 A-P early CRMs and 16 D-V CRMs. The intensities of all factors were placed on a similar scale by normalizing the data such that the intensity score of the most highly bound region in the genome for each factor is set to 10.
Figure 13
Heat maps showing two measures for factor binding specificity in blastoderm embryos. (a, b) Each row shows the GSC z score of the likelihood that the overlap between a cohort of ChIP/chip peaks for one factor and regions bound each factor in turn occurs by chance. (c, d) Each row shows the Pearson correlation coefficients between the intensity scores of a cohort of 300 peaks for a factor and the intensity scores of the equivalent 500-bp windows at the same genomic locations for each of the other factors in turn. (a, c) Results for the most highly bound 300 peaks (1-300). (b, d) Results for the second most highly bound cohort of 300 peaks (301-600). Note that the 1% FDR threshold does not lie within ranks 1 to 600 for 17 of the 21 factors (Table 2), and, thus, for these proteins the differences observed between the 1-300 and the 301-600 cohorts are not attributable to false positives.
Figure 14
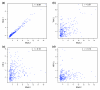
Scatter plots showing the correlation between 500-bp window scores. The 500-bp peak window scores for the top 300 regions detected by the SNA 2 antibody (x-axis) are compared against the score of the equivalent 500-bp windows detected in another Chip/chip experiment (y-axis) at the same genomic locations. The comparison is made against ChIP/chip data from experiments using the (a) SNA 1, (b) TWI 2, (c) Kruppel (KR) 2, and (d) HRY 2 antibodies. The Pearson correlation coefficients (r) for each comparison are shown in the top right of each panel.
Figure 15
Heat map showing GO terms enriched in genes closest to regions bound by each factor. The seven most highly enriched GO terms associated with the closest genes to the 300 most highly bound peaks were determined for each of the 21 factors and the non-redundant set of all such terms identified. Each row shows the enrichment of each of these GO terms for one factor expressed as a normalized z score. The columns (GO terms) were arranged into three groups based on which of the three major regulatory classes of factor the GO terms are most enriched in, and are ranked from left to right based on the degree of this relative enrichment. (a) Results for the most highly bound 300 peaks (1-300); (b) results for the second most highly bound cohort of 300 peaks (301-600).
Similar articles
- Transcription factors bind thousands of active and inactive regions in the Drosophila blastoderm.
Li XY, MacArthur S, Bourgon R, Nix D, Pollard DA, Iyer VN, Hechmer A, Simirenko L, Stapleton M, Luengo Hendriks CL, Chu HC, Ogawa N, Inwood W, Sementchenko V, Beaton A, Weiszmann R, Celniker SE, Knowles DW, Gingeras T, Speed TP, Eisen MB, Biggin MD. Li XY, et al. PLoS Biol. 2008 Feb;6(2):e27. doi: 10.1371/journal.pbio.0060027. PLoS Biol. 2008. PMID: 18271625 Free PMC article. - DNA regions bound at low occupancy by transcription factors do not drive patterned reporter gene expression in Drosophila.
Fisher WW, Li JJ, Hammonds AS, Brown JB, Pfeiffer BD, Weiszmann R, MacArthur S, Thomas S, Stamatoyannopoulos JA, Eisen MB, Bickel PJ, Biggin MD, Celniker SE. Fisher WW, et al. Proc Natl Acad Sci U S A. 2012 Dec 26;109(52):21330-5. doi: 10.1073/pnas.1209589110. Epub 2012 Dec 10. Proc Natl Acad Sci U S A. 2012. PMID: 23236164 Free PMC article. - Dynamic reprogramming of chromatin accessibility during Drosophila embryo development.
Thomas S, Li XY, Sabo PJ, Sandstrom R, Thurman RE, Canfield TK, Giste E, Fisher W, Hammonds A, Celniker SE, Biggin MD, Stamatoyannopoulos JA. Thomas S, et al. Genome Biol. 2011;12(5):R43. doi: 10.1186/gb-2011-12-5-r43. Epub 2011 May 11. Genome Biol. 2011. PMID: 21569360 Free PMC article. - Regulation of segmentation and segmental identity by Drosophila homeoproteins: the role of DNA binding in functional activity and specificity.
Biggin MD, McGinnis W. Biggin MD, et al. Development. 1997 Nov;124(22):4425-33. doi: 10.1242/dev.124.22.4425. Development. 1997. PMID: 9409661 Review. - Building quantitative, three-dimensional atlases of gene expression and morphology at cellular resolution.
Knowles DW, Biggin MD. Knowles DW, et al. Wiley Interdiscip Rev Dev Biol. 2013 Nov-Dec;2(6):767-79. doi: 10.1002/wdev.107. Epub 2013 Feb 4. Wiley Interdiscip Rev Dev Biol. 2013. PMID: 24123936 Free PMC article. Review.
Cited by
- Identifying targets of the Sox domain protein Dichaete in the Drosophila CNS via targeted expression of dominant negative proteins.
Shen SP, Aleksic J, Russell S. Shen SP, et al. BMC Dev Biol. 2013 Jan 5;13:1. doi: 10.1186/1471-213X-13-1. BMC Dev Biol. 2013. PMID: 23289785 Free PMC article. - Integrative analysis of C. elegans modENCODE ChIP-seq data sets to infer gene regulatory interactions.
Van Nostrand EL, Kim SK. Van Nostrand EL, et al. Genome Res. 2013 Jun;23(6):941-53. doi: 10.1101/gr.152876.112. Epub 2013 Mar 26. Genome Res. 2013. PMID: 23531767 Free PMC article. - Effects of sequence variation on differential allelic transcription factor occupancy and gene expression.
Reddy TE, Gertz J, Pauli F, Kucera KS, Varley KE, Newberry KM, Marinov GK, Mortazavi A, Williams BA, Song L, Crawford GE, Wold B, Willard HF, Myers RM. Reddy TE, et al. Genome Res. 2012 May;22(5):860-9. doi: 10.1101/gr.131201.111. Epub 2012 Feb 2. Genome Res. 2012. PMID: 22300769 Free PMC article. - Disentangling the many layers of eukaryotic transcriptional regulation.
Lelli KM, Slattery M, Mann RS. Lelli KM, et al. Annu Rev Genet. 2012;46:43-68. doi: 10.1146/annurev-genet-110711-155437. Epub 2012 Aug 28. Annu Rev Genet. 2012. PMID: 22934649 Free PMC article. Review. - Identification of transcription factor high accumulation DNA zones.
Cascianelli S, Ceddia G, Marchesi A, Masseroli M. Cascianelli S, et al. BMC Bioinformatics. 2023 Oct 20;24(1):395. doi: 10.1186/s12859-023-05528-1. BMC Bioinformatics. 2023. PMID: 37864168 Free PMC article.
References
- Davidson EH. Genomic Regulatory Systems - Development and Evolution. San Diego: Academic Press; 2001.
- Ptashne M, Gann A. Genes and Signals. New York: Cold Spring Harbor Press; 2002.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases