YY1's role in DNA methylation of Peg3 and Xist - PubMed (original) (raw)
YY1's role in DNA methylation of Peg3 and Xist
Jeong Do Kim et al. Nucleic Acids Res. 2009 Sep.
Abstract
Unusual clusters of YY1 binding sites are located within several differentially methylated regions (DMRs), including Xist, Nespas and Peg3, which all become methylated during oogenesis. In this study, we performed conditional YY1 knockdown (KD) to investigate YY1's roles in DNA methylation of these DMRs. Reduced levels of YY1 during spermatogenesis did not cause any major change in these DMRs although the same YY1 KD caused hypermethylation in these DMRs among a subset of aged mice. However, YY1 KD during oogenesis resulted in the loss of DNA methylation on Peg3 and Xist, but there were no changes on Nespas and H19. Continued YY1 KD from oogenesis to the blastocyst stage caused further loss in DNA methylation on Peg3. Consequently, high incidents of lethality were observed among embryos that had experienced the reduced levels of YY1 protein. Overall, the current study suggests that YY1 likely plays a role in the de novo DNA methylation of the DMRs of Peg3 and Xist during oogenesis and also in the maintenance of unmethylation status of these DMRs during spermatogenesis.
Figures
Figure 1.
Overall Strategy of conditional YY1 KD. (A) Schematic representation of the pSico-YY1 shRNA vector. The U6 promoter is separated from the downstream YY1-shRNA sequence by the EGFP expression cassette. Cre recombinase, available as a transgenic allele, recognizes the two TATA-loxP sites and joins these two split regions, resulting in the production of the YY1 shRNA sequence, which leads to KD. The sequence in blue is the leader sequence of the U6 RNA gene, which has been added to increase the stability of the YY1 shRNA, and the sequence in red is the sequence of the YY1 shRNA. (B) Conditional KD scheme for gametogenesis. The pSico-YY1 transgenic line was crossed with one of two germ cell-specific Cre mice: Protamine-Cre for spermatogenesis and Zp3-Cre for oogenesis. The resulting F1 mice were used for testing potential effects of YY1 KD during either spermatogenesis or oogenesis.
Figure 2.
YY1 KD effects during spermatogenesis. Sperm DNA was isolated individually from the F1 mice (pSico-YY1, Pro-Cre) and the littermate controls (+, Pro-Cre). The individual F1 mice are indicated by different numbers, while the control littermates are indicated by numbers followed by C (1C, 5C, 8C and 14C). (A) Individual DNA samples from the 3-month-old set were treated with the bisulfite conversion method, followed by the restriction-enzyme-based method—COBRA. Digestion and lack of digestion by TaqI and ClaI indicate the methylated (M) and unmethylated (U) status, respectively, of a given CpG site in the original DNA as shown in the digestion of Peg3, Xist, Tsix and H19. In contrast, this is opposite for the HphI digestion, which recognizes TpG as shown in the digestion of Nespas. Digestion by this enzyme indicates the unmethylated status of a CpG site, whereas lack of digestion indicates methylation. (B) A subset of the 6-month-old mice is also shown in the same manner as the above. The results for H19 (8#C and 9) are not available due to the unsuccessful PCR amplification.
Figure 3.
YY1 KD effects during oogenesis. About 400 eggs at MII phase were isolated from each of the following two types of female mice: the KD mice (pScio-YY1, Zp3-Cre) and the control (Con) littermates (+, Zp3-Cre). Genomic DNA from these two types of mice was first treated by bisulfite conversion, followed by restriction enzyme digestion (A) and sequencing (B). The PCR product from the H19-ICR was digested by ClaI, while the remaining products from the DMRs of Nespas, Peg3 and Xist were digested by TaqI. The results derived from sequencing of individual PCR products were summarized in the following manner. Each circle represents one CpG site: closed and open ones indicate methylated and unmethylated states, respectively, of the analyzed CpG sites. Each row represents one clone derived from the original PCR product. The methylation levels (%) of each DMR were calculated through dividing the number of methylated CpGs by the total number of surveyed CpG sites. We monitored and compared the purity, number and morphology of the isolated egg samples, and a representative image of the isolated egg is also shown (C).
Figure 4.
YY1 KD effects during early embryogenesis. Blastocysts were isolated through crossing the female F1 mice (pSico-YY1, Zp3-Cre) with littermate breeders. The isolated blastocysts were categorized based on their genotypes and genders. The DNA from each category of blastocysts was first treated with the bisulfite conversion, followed by the restriction digestion-based COBRA (A) and sequencing (B). The digested PCR products for H19, Nespas and Peg3 are derived from male blastocysts, while Xist is shown in four categories due to the fact that X chromosome number varies depending on gender. The PCR products were digested by the following restriction enzymes: H19 was digested by ClaI, while Nespas, Peg3 and Xist were digested by TaqI. The sequencing results for Xist are from female blastocysts (Con and KD).
Figure 5.
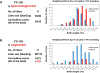
Developmental potential of the gametes with YY1 KD. The germ cells with YY1 KD were also tested through breeding the male F1 mice (pScio-YY1, Pro-Cre) (A) and the female F1 mice (pSico-YY1, Zp3-Cre) (B) with their littermates. These two breeding experiments were summarized as shown on the left. The two graphs on the right represent the birth weight profiles derived from the 1-day-old neonates of these two breeding experiments. The neonates were analyzed in terms of their birth weights. Each mouse was first categorized according to its genotype [carrier (red) versus noncarrier (blue)]. The relative weight percentile for each mouse was calculated through dividing its weight by the averaged value for its litter. This relative weight value was used to further classify each mouse into different categories ranging from 65% to 125% as shown on the _x_-axis. The total number of mice in each weight category is plotted as a value on the _y_-axis.
Similar articles
- In vivo YY1 knockdown effects on genomic imprinting.
Kim J, Kim JD. Kim J, et al. Hum Mol Genet. 2008 Feb 1;17(3):391-401. doi: 10.1093/hmg/ddm316. Epub 2007 Oct 31. Hum Mol Genet. 2008. PMID: 17977899 - YY1's role in the Peg3 imprinted domain.
He H, Ye A, Perera BPU, Kim J. He H, et al. Sci Rep. 2017 Jul 25;7(1):6427. doi: 10.1038/s41598-017-06817-5. Sci Rep. 2017. PMID: 28743993 Free PMC article. - YY1 as a controlling factor for the Peg3 and Gnas imprinted domains.
Kim JD, Hinz AK, Choo JH, Stubbs L, Kim J. Kim JD, et al. Genomics. 2007 Feb;89(2):262-9. doi: 10.1016/j.ygeno.2006.09.009. Epub 2006 Oct 25. Genomics. 2007. PMID: 17067777 Free PMC article. - Physical Interaction of Human Yin Yang 1 Protein with DNA.
Figiel M, Górecki A. Figiel M, et al. Crit Rev Oncog. 2017;22(1-2):75-97. doi: 10.1615/CritRevOncog.2017020976. Crit Rev Oncog. 2017. PMID: 29604938 Review. - The Yin and Yang of YY1 in tumor growth and suppression.
Khachigian LM. Khachigian LM. Int J Cancer. 2018 Aug 1;143(3):460-465. doi: 10.1002/ijc.31255. Epub 2018 Jan 31. Int J Cancer. 2018. PMID: 29322514 Review.
Cited by
- Oncogenic potential of yin yang 1 mediated through control of imprinted genes.
Thiaville MM, Kim J. Thiaville MM, et al. Crit Rev Oncog. 2011;16(3-4):199-209. doi: 10.1615/critrevoncog.v16.i3-4.40. Crit Rev Oncog. 2011. PMID: 22248054 Free PMC article. Review. - Epigenetic polymorphism and the stochastic formation of differentially methylated regions in normal and cancerous tissues.
Landan G, Cohen NM, Mukamel Z, Bar A, Molchadsky A, Brosh R, Horn-Saban S, Zalcenstein DA, Goldfinger N, Zundelevich A, Gal-Yam EN, Rotter V, Tanay A. Landan G, et al. Nat Genet. 2012 Nov;44(11):1207-14. doi: 10.1038/ng.2442. Epub 2012 Oct 14. Nat Genet. 2012. PMID: 23064413 - Identification of an evolutionarily conserved cis-regulatory element controlling the Peg3 imprinted domain.
Thiaville MM, Kim H, Frey WD, Kim J. Thiaville MM, et al. PLoS One. 2013 Sep 10;8(9):e75417. doi: 10.1371/journal.pone.0075417. eCollection 2013. PLoS One. 2013. PMID: 24040411 Free PMC article. - Identification and Functional Analysis of E3 Ubiquitin Ligase g2e3 in Chinese Tongue Sole, Cynoglossus semilaevis.
Cui Z, Luo J, Cheng F, Xu W, Wang J, Lin M, Sun Y, Chen S. Cui Z, et al. Animals (Basel). 2024 Sep 5;14(17):2579. doi: 10.3390/ani14172579. Animals (Basel). 2024. PMID: 39272364 Free PMC article. - Retrotransposon-derived promoter of Mammalian Aebp2.
Kim H, Bakshi A, Kim J. Kim H, et al. PLoS One. 2015 Apr 27;10(4):e0126966. doi: 10.1371/journal.pone.0126966. eCollection 2015. PLoS One. 2015. PMID: 25915901 Free PMC article.
References
- Edwards CA, Ferguson-Smith AC. Mechanisms regulating imprinted genes in clusters. Curr. Opin. Cell Biol. 2007;19:281–289. - PubMed
- Reik W. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature. 2007;447:425–432. - PubMed
- Sasaki H, Matsui Y. Epigenetic events in mammalian germ-cell development: reprogramming and beyond. Nat. Rev. Genet. 2008;9:29–140. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases