X11alpha haploinsufficiency enhances Abeta amyloid deposition in Alzheimer's disease transgenic mice - PubMed (original) (raw)
X11alpha haploinsufficiency enhances Abeta amyloid deposition in Alzheimer's disease transgenic mice
Inderjeet Saluja et al. Neurobiol Dis. 2009 Oct.
Abstract
The neuronal adaptor protein X11alpha/mint-1/APBA-1 binds to the cytoplasmic domain of the amyloid precursor protein (APP) to modulate its trafficking and metabolism. We investigated the consequences of reducing X11alpha in a mouse model of Alzheimer's disease (AD). We crossed hAPPswe/PS-1DeltaE9 transgenic (AD tg) mice with X11alpha heterozygous knockout mice in which X11alpha expression is reduced by approximately 50%. The APP C-terminal fragments C99 and C83, as well as soluble Abeta40 and Abeta42, were increased significantly in brain of X11alpha haploinsufficient mice. Abeta/amyloid plaque burden also increased significantly in the hippocampus and cortex of one year old AD tg/X11alpha (+/-) mice compared to AD tg mice. In contrast, the levels of sAPPalpha and sAPPbeta were not altered significantly in AD tg/X11alpha (+/-) mice. The increased neuropathological indices of AD in mice expressing reduced X11alpha suggest a normal suppressor role for X11alpha on CNS Abeta/amyloid deposition.
Figures
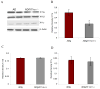
Fig. 1. Reduced X11α protein in ADtg/X11α(+/−) mouse brain without changes in other X11 family proteins
(A) Immunoblot analysis of X11α in brain homogenates confirmed a reduced level of X11α in AD tg/X11α +/− mice (Lanes 3 and 4) compared to AD tg mice (Lanes 1 and 2), while there was no change in the levels of X11β and X11γ. (B) Semi-quantitative analyses of immunoblots revealed that the X11α band density in brains from AD tg/X11α +/− mice was significantly less (54 %) than in AD tg mice. (C–D) Semi-quantitative analyses of the band density for X11β and X11γ revealed no significant change in the levels of either protein. Error bars indicate S.E.M. (n = 6), and statistically significant differences are indicated (*p < 0.01)
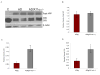
Fig. 2. Increased C-terminal fragments in AD tg/X11α+/− mice
(A) Immunoblot analysis of APP and the C-terminal fragments C99 (CTFβ) and C83 (CTFα). Proteins (65 μg) from mouse brain homogenates were detected by immunoblot with an antibody to the N-terminus of APP (for detecting full length APP), to the C-terminus of APP (for detecting CTFs), or to β-actin. Protein from cell lysates transfected with C99 (Lane 1) was run as a positive control for detecting the C99 fragment in brain homogenate (B, C, D). Band intensities of total APP, C83 and C99 from AD tg and AD tg/X11α +/− mice were quantified and normalized to the levels of β-actin. X11α haploinsufficiency significantly increased the level of C83 (CTFα) (C) and C99 (CTFβ) (D) without altering the level of total APP (B). Error bars indicate S.E.M. (n = 6), and statistically significant differences are indicated (**p < 0.01 and *p <0.05)
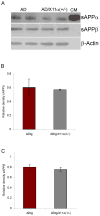
Fig. 3. Levels of sAPPα and sAPPβ fragments in AD tg/X11α (+/−) mice
Immunoblot analysis of sAPPα and sAPPβ fragments is shown. Proteins (60 μg) from mouse brain homogenates were detected by immunoblot with antibodies to sAPPα, sAPPβ or to β-actin. Conditioned medium (CM) from a CHO (Chinese hamster ovary) cell line stably transfected with APPswe and PS-1ΔE9 mutation was used as a positive control to verify the specificity of the secreted product. (A) Band intensities were quantified and normalized to levels of β-actin. X11α haploinsufficiency does not lead to a statistically significant change in the level of (B) sAPPα and (C) sAPPβ. Error bars indicate S.E.M. (n = 4).
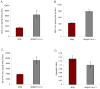
Fig. 4. Aβ levels in brain are increased by X11α reduction
Measurement of soluble Aβ levels in 12 month old mouse brain homogenates revealed a significant increase in Aβ40 (A), Aβ42 (B), and total Aβ (C) levels in ADtg mice on an X11α haploinsufficient background (n = 4; * indicates p < 0.01). The Aβ42/Aβ40 ratio (D) was not significantly altered.
Fig. 5. Aβ/amyloid plaque density is increased by X11α reduction
(A) Representative silver-stained sections from the hippocampus and cortex of AD tg mice or of AD tg/X11α haploinsufficient mice revealed a higher plaque density in AD tg mice on a X11α +/− background. Higher magnification images in lower four panels represent insets from the top two panels. Scale bars 150 μ (top panels) and 50 μ (bottom four panels). (B–C) Increased plaque density in hippocampus of ADtg/X11α depleted mice. Stereological counts of amyloid plaque density were obtained from sections from cortex and hippocampus of AD tg versus ADtg/X11α +/− mice at 12 months of age. The total number of plaques in the cortex (B) and hippocampus (C) were calculated for both groups and expressed as total counts per section. The error bars indicate S.E.M (n = 4), and statistically significant differences are indicated (*p < 0.05).
Similar articles
- Deletion of Mint proteins decreases amyloid production in transgenic mouse models of Alzheimer's disease.
Ho A, Liu X, Südhof TC. Ho A, et al. J Neurosci. 2008 Dec 31;28(53):14392-400. doi: 10.1523/JNEUROSCI.2481-08.2008. J Neurosci. 2008. PMID: 19118172 Free PMC article. - Overexpression of Ubiquilin-1 Alleviates Alzheimer's Disease-Caused Cognitive and Motor Deficits and Reduces Amyloid-β Accumulation in Mice.
Adegoke OO, Qiao F, Liu Y, Longley K, Feng S, Wang H. Adegoke OO, et al. J Alzheimers Dis. 2017;59(2):575-590. doi: 10.3233/JAD-170173. J Alzheimers Dis. 2017. PMID: 28598849 Free PMC article. - X11alpha impairs gamma- but not beta-cleavage of amyloid precursor protein.
King GD, Cherian K, Turner RS. King GD, et al. J Neurochem. 2004 Feb;88(4):971-82. doi: 10.1046/j.1471-4159.2003.02234.x. J Neurochem. 2004. PMID: 14756819 - The X11/Mint family of adaptor proteins.
Rogelj B, Mitchell JC, Miller CC, McLoughlin DM. Rogelj B, et al. Brain Res Rev. 2006 Sep;52(2):305-15. doi: 10.1016/j.brainresrev.2006.04.005. Epub 2006 Jun 9. Brain Res Rev. 2006. PMID: 16764936 Review. - Mechanisms of AD neurodegeneration may be independent of Aβ and its derivatives.
Robakis NK. Robakis NK. Neurobiol Aging. 2011 Mar;32(3):372-9. doi: 10.1016/j.neurobiolaging.2010.05.022. Epub 2010 Jul 1. Neurobiol Aging. 2011. PMID: 20594619 Free PMC article. Review.
Cited by
- Interrogation and validation of the interactome of neuronal Munc18-interacting Mint proteins with AlphaFold2.
Weeratunga S, Gormal RS, Liu M, Eldershaw D, Livingstone EK, Malapaka A, Wallis TP, Bademosi AT, Jiang A, Healy MD, Meunier FA, Collins BM. Weeratunga S, et al. J Biol Chem. 2024 Jan;300(1):105541. doi: 10.1016/j.jbc.2023.105541. Epub 2023 Dec 9. J Biol Chem. 2024. PMID: 38072052 Free PMC article. - GULP1/CED-6 ameliorates amyloid-β toxicity in a Drosophila model of Alzheimer's disease.
Vivien Chiu WY, Koon AC, Ki Ngo JC, Edwin Chan HY, Lau KF. Vivien Chiu WY, et al. Oncotarget. 2017 Aug 8;8(59):99274-99283. doi: 10.18632/oncotarget.20062. eCollection 2017 Nov 21. Oncotarget. 2017. PMID: 29245900 Free PMC article. - Amyloid precursor protein processing and Alzheimer's disease.
O'Brien RJ, Wong PC. O'Brien RJ, et al. Annu Rev Neurosci. 2011;34:185-204. doi: 10.1146/annurev-neuro-061010-113613. Annu Rev Neurosci. 2011. PMID: 21456963 Free PMC article. Review. - Amyloid precursor protein and its interacting proteins in neurodevelopment.
Chau DD, Ng LL, Zhai Y, Lau KF. Chau DD, et al. Biochem Soc Trans. 2023 Aug 31;51(4):1647-1659. doi: 10.1042/BST20221527. Biochem Soc Trans. 2023. PMID: 37387352 Free PMC article. Review. - Activation of liver X receptor decreases BACE1 expression and activity by reducing membrane cholesterol levels.
Cui W, Sun Y, Wang Z, Xu C, Xu L, Wang F, Chen Z, Peng Y, Li R. Cui W, et al. Neurochem Res. 2011 Oct;36(10):1910-21. doi: 10.1007/s11064-011-0513-3. Epub 2011 Jun 1. Neurochem Res. 2011. PMID: 21630010
References
- Araki Y, Tomita S, Yamaguchi H, Miyagi N, Sumioka A, Kirino Y, Suzuki T. Novel cadherin-related membrane proteins, alcadeins, enhance the X11-like protein-mediated stabilization of amyloid β-protein precursor metabolism. J Biol Chem. 2003;278:49448–58. - PubMed
- Borg JP, Straight SW, Kaech SM, de Taddéo-Borg M, Kroon DE, Karnak D, Turner RS, Kim SK, Margolis B. Identification of an evolutionarily conserved heterotrimeric protein complex involved in protein targeting. J Biol Chem. 1998a;273:31633–6. - PubMed
- Borg JP, Yang Y, De Taddéo-Borg M, Margolis B, Turner RS. The X11α protein slows cellular amyloid precursor protein processing and reduces Aβ40 and Aβ42 secretion. J Biol Chem. 1998b;273:14761–6. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous